
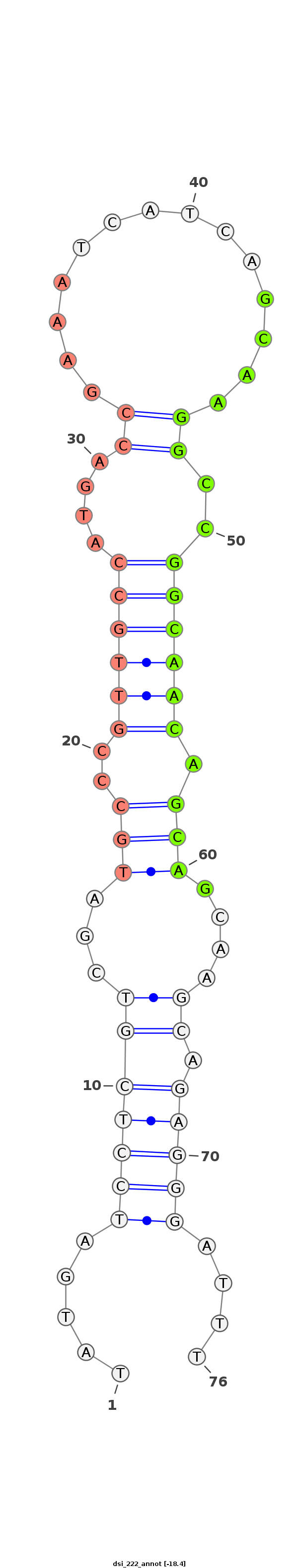


ID:dsi_222 |
Coordinate:2R:11376756-11376802 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -17.8 | -17.8 | -17.5 |
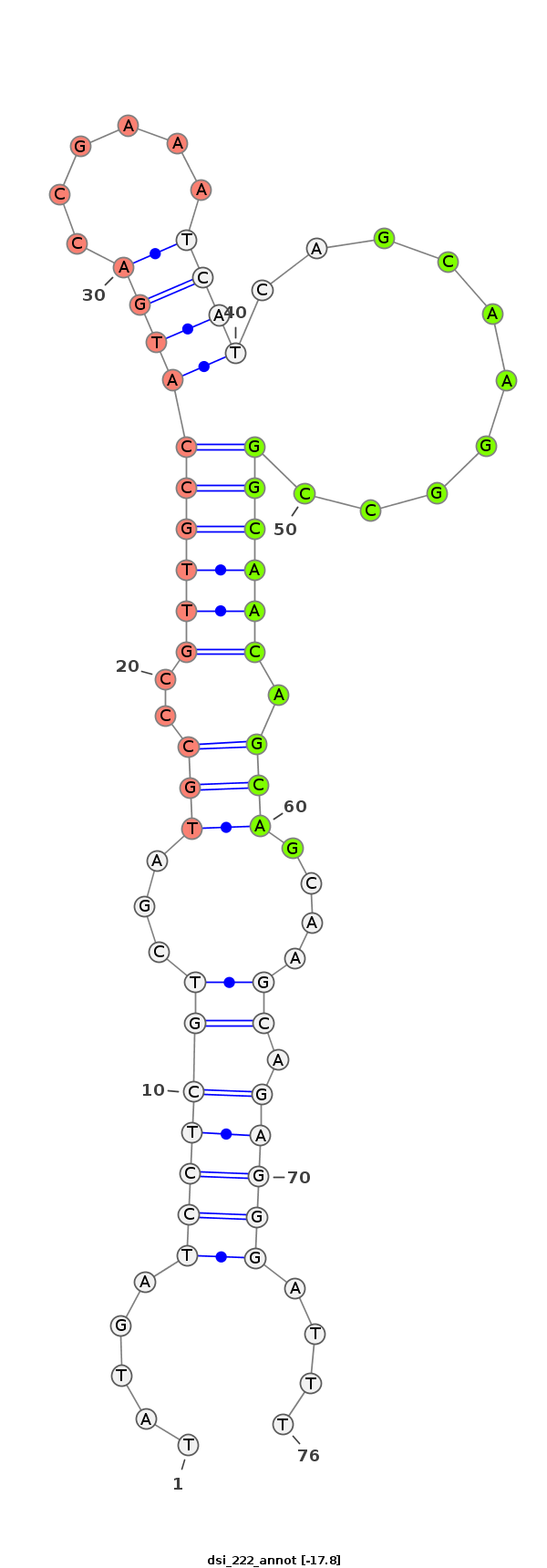 |
 |
 |
intron [Dsim\GD25516-in]
No Repeatable elements found
|
GCGGCCAGGATCCGGGTCCACATCATGGTTCAGGATATGATCCTCGTCGATGCCCGTTGCCATGACCGAAATCATCAGCAAGGCCGGCAACAGCAGCAAGCAGAGGGATTTGAACCAAACCATCTTACAAACCAAAAGCAACTGAAG
***********************************.....(((((((...(((..((((((....((..............))..)))))).)))....)).)))))....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
O002 Head |
M053 Female-body |
|---|---|---|---|---|---|---|---|---|
| .............................................................................GCAAGGCCGGCAACAGCAG................................................... | 19 | 0 | 1 | 12.00 | 12 | 9 | 3 | 0 |
| .............................................................................GCAAGGCCGGCAACAGCAGCA................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................................................................GCAAGCAGAGGGATTTGAACCAAAC........................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ......................................GATCCTCGTCGATGCCCGTTGC....................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................AGCAAGGCCGGCAACAGCAG................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................TGCCCGTTGCCATGACCGAAA............................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ............................................................................................................TTTGAACCAAACGTTATTACAAA................ | 23 | 3 | 2 | 1.00 | 2 | 0 | 0 | 0 |
| .......................................................GCTGCCATGACCGAA............................................................................. | 15 | 1 | 4 | 0.50 | 2 | 0 | 0 | 0 |
| ....................................GTGATCCTCGTCGATGCCG............................................................................................ | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 |
| ..................CAAATCATGGTTAAGGAT............................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 |
| ................................................GATTCCCGTTGCCTTGACTG............................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 |
| .........................................................TGCCATGTCCGAAAT........................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 |
| .........................................................................................ACAGAAGCAAGCAGA........................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 |
|
CTTCAGTTGCTTTTGGTTTGTAAGATGGTTTGGTTCAAATCCCTCTGCTTGCTGCTGTTGCCGGCCTTGCTGATGATTTCGGTCATGGCAACGGGCATCGACGAGGATCATATCCTGAACCATGATGTGGACCCGGATCCTGGCCGC
************************************....(((((.((....(((.((((((..((..............))....))))))..)))...))))))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
|---|---|---|---|---|---|---|
| .........................................................................................................................ATGATGTAGACCCGAAACCT...... | 20 | 3 | 1 | 8.00 | 8 | 1 |
| ........................................................................................................................CATGATGTAGACCCGAAACCT...... | 21 | 3 | 1 | 7.00 | 7 | 5 |
| ........................................................................................................................CATGATGTAGACCCGAAACC....... | 20 | 3 | 1 | 5.00 | 5 | 3 |
| ................................................................................................ATCGACGAGGATCATATCCTGAACC.......................... | 25 | 0 | 1 | 2.00 | 2 | 2 |
| ................................................................................................................TCCGGAACCAGGATGTGGA................ | 19 | 2 | 3 | 1.67 | 5 | 0 |
| ...................................................................TGCTGATGATTTCGGTCAT............................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 |
| ................................................................................................ATCGACGAGGATCATATCCTGAAC........................... | 24 | 0 | 1 | 1.00 | 1 | 1 |
| ...............................GGTTCAAATCCCTCCGCTCGT............................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 |
| ................................GTTCAAATCCCTCCGCT.................................................................................................. | 17 | 1 | 1 | 1.00 | 1 | 0 |
| .................................................................................................................CCGGAACCAGGATGTGGA................ | 18 | 2 | 8 | 0.88 | 7 | 0 |
| ....................................................TGCTGTTGCCGGCCTTGTCGGT......................................................................... | 22 | 3 | 3 | 0.67 | 2 | 0 |
| .................................................................................................................CCGGAACCAGGATGTGGATCC............. | 21 | 3 | 3 | 0.67 | 2 | 0 |
| .........................................................................................................................ATGATGTAGACCCGAAACC....... | 19 | 3 | 9 | 0.56 | 5 | 2 |
| ........................................................................................................................CATGATGTAGACCCGAAAC........ | 19 | 3 | 10 | 0.50 | 5 | 3 |
| ..........................................................................................................................TGATGTAGACCCGAAACCT...... | 19 | 3 | 8 | 0.38 | 3 | 2 |
| .................................TTCAAATCCCTCTGCTCGT............................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 |
| ....................................................................................................................GTATCATGATGTAGACCCG............ | 19 | 3 | 6 | 0.33 | 2 | 2 |
| ................................GTTCGAATCCCTCTGC................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 |
| ................................GTTCGAATCCCTTCGCTTGCT.............................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 |
| ................................GTTCAAATCCCTCCGCTCGT............................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 |
| ..............................................................................................................................GTTGACCCGGTTCCCGGCC.. | 19 | 3 | 17 | 0.12 | 2 | 0 |
| ................................................................................................................TCCGGAACCAGGATGTG.................. | 17 | 2 | 19 | 0.11 | 2 | 0 |
| .................................................................................................................CCTGAACCAGGACGTGGA................ | 18 | 2 | 13 | 0.08 | 1 | 0 |
| .................TTGTGAGATAGTTTGGTT................................................................................................................ | 18 | 2 | 13 | 0.08 | 1 | 0 |
| ..................................................................................................................CGGAACCATGATGTGGATT.............. | 19 | 3 | 16 | 0.06 | 1 | 0 |
| ....................................................................................................................GAACCAGGATGTGGATCCT............ | 19 | 3 | 17 | 0.06 | 1 | 0 |
| ................................................................................................................TCCGGAACCAGAATGTGGA................ | 19 | 3 | 17 | 0.06 | 1 | 0 |
| ................................GTTCAAATCCTTCCGCTCG................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 |
| ...............................GGTTCAAATCCGTCCGCC.................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 |
| ......................................................................................................................ATCATGATGTAGACCCGAA.......... | 19 | 3 | 20 | 0.05 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 2R:11376706-11376852 - | dsi_222 | GCGGCC-AGGATCCGGGTCCACATCATGGTTCAGGATATGATCCTCGTCGATGCCCGTTGCCATGACCGAAATCATCAGCAAGGCCGGCAACAGCAGCA------AGCAGAGGGATTTGA---ACCAAACCATCTTACAAACCAAAAGCAACTGAAG----- |
| droSec2 | scaffold_1:10147455-10147601 - | GCGGCC-GGGATCCGGGTCCACATCATGGTTCAGGATATGATCCTCGTCGATGCCCGTTGCCATGAGCGAATTCATCAGTAAGGCCGGCAGCAGCAGCA------AGCAGAGGGATTTGA---ACCAAACCATCCTACAAACCAAAAGCAACTGAAG----- | |
| dm3 | chr2R:12641988-12642134 - | GCGGCC-CGGATCCGGGTCCACATCATGGTTCAGGATATGATCCTCGTCGATGCCAGTTGTCATGACCGAATCCATCAGCAAGGCCGGCAGCAGCAGAA------AGCAGAGGGAATTGA---ACCAAACCATCATGCAAAACAAAAGCAACTGAAG----- | |
| droEre2 | scaffold_4845:13209605-13209748 + | GCGACC-CGGATCCGGGTCCACATCATGGTTAACGACATGATCCTCGTC---GTCAGTTGACGTGACCGAATTCATCAGTAAGGCCGGCAGCAGCAGCA------AGCAGAGAGATTTTA---ACCGAGCCATCCTACAAATCAAAAGCAACTGAAG----- | |
| droYak3 | 2R:16335835-16335978 + | GCGTCC-CGGATCCGGGTCCACATCATGGTCTAGGATATGATCCTCGTC---GTCAGTTGACGTGACCGAAATCATCAGTAAGGCCGGCAGCAGCAGCA------AGCAGAGTGATTTAA---ACCAAACCATCCTACAAATCAAAAGCAACTGAAG----- | |
| droEug1 | scf7180000409672:2505304-2505456 + | GCGACC-CGGATCCGGGTCCACTTCATGGTTTATAACTGTTTGCTCAGGAATGCCAGTTGCCCGAACCGAAATCATTAATAAAGCCGGCAGCAGCAGCAGCAGCCAACAGAGAGACTTTA---ACTTAATCATCCTAAGAATCAAAAGCAACTGAAG----- | |
| droBia1 | scf7180000301506:2039974-2040120 - | CCGACC-CGGATCCGGGTCTTCATAATGGTTCAGTACCGTTTCCTCTGCAAAGCCAGTTGACATAGCCGAATTTATAAATAAAACTGGTAGTACCAGCC------AAGAAAGAGACTTGA---TCCAAACCATCCTATAAGCCAAACGCAACTGAAG----- | |
| droTak1 | scf7180000414504:27279-27425 - | ACGACT-CGGATCCGGGTCCACTTCGTGGTTAAGAACAGTTTCCTCCGCAAAGCCAGTTGTCATCACGGAATTCACAAATAATGCCGGCAGCAGCAGCA------AAAAAAGAGACTTTA---ACCAAATCATCCTGCAAACCAGAGGCAACTGAAA----- | |
| droEle1 | scf7180000491214:1741711-1741854 + | ACGATC-CGGATCCGGGTCCACCTCATGGTTGATAATGTAATTTC---CAAAGCCAGTTGACATCACTGTTCTTATTAAAAACGCTGGCAACAGCAGCA------AACAGAGAAGCTTAA---ACCAAATCATTCTTAAAGCCAAATGCAACTGAGG----- | |
| droRho1 | scf7180000780084:710438-710584 + | GCGACT-CGGATCCGGGTCCACCTCATGGTGGAGAACTTGTTCCTCTGCAAAGCCTGTTGCCATCACCGATTTCAACAATAACGCCGGCAGCAGCAGCA------AACAGAGCGACGTTA---ACCAAAACATCCTAAAAACCTGAAGCAACTGAGG----- | |
| droFic1 | scf7180000453851:855598-855744 - | GCGACC-CGGATCCGGGTCACCTTCATGGCTCAAAATGTGTTCCTCTGCAAAGCCAGTTGTCATCAAAGAAATCATGAATACAGCTGGCAGCAGCAGCA------AATAGAGAAACTTCA---ACTCGATCATCGTACGAACAAAAGGCAACTGAAG----- | |
| droKik1 | scf7180000302470:1050093-1050239 - | ACGATC-CGGATCCGGGTCCACCTTATGGTGTAGAACAGTCTCAT---CAAAGCTAGTAACCATCACCATTTTCATGAACAAAGCTGCTGGCAGAAGCA------AAGAAAGGGAGAATAGCAACCGAGCCATTATAGAAACCAAAAGCAACTGGAG----- | |
| droAna3 | scaffold_13266:13949661-13949804 + | CCGAGT-CGGATCCGGGTTTTTATCGTGGCCCAAAATTGTTTCCAAAGGCAAGCTTTCGGCCATCAATGCTGGAAAGTATATAGCTGGCAGCAGCAGGA-------GC--AGAAAGCTGG---AGCACATCATCCTCCAAGTCGGTGCAAACTCAAA----- | |
| droBip1 | scf7180000396759:2187713-2187861 - | CCGATT-CGGATCCGGGTCTAGTTTGTGGCTCAAAACCGTTTCCGATGACCAGCTGACGGACATCAGTGTTGGGATGTATATAGCTGGCAGCAGCAGGA-------GCA--GAAAACTGG---AGCACATCATCCTCCAAGTTGGATCAAGCTCAAACTAAT | |
| dp5 | 3:19069459-19069581 + | GCGATC-CGGGTCCGGGATTGCGTCATGAACCAACACATCGCTC------------ATTT------CCATGACCACAACGAGAGCTGGCAGCAGCAAAA-------ACA-----GACTCA---ACCGAATCATTCGGCGTTTCGTGACTAACTGAAA----- | |
| droPer2 | scaffold_37:72247-72369 + | GCGATC-CGGGTCCGGGATTGCGTCATGAACCAACACATCGCTC------------ATTT------CCATGACCACAACGAGAGCTGGCAGCAGCAGAA-------ACA-----GACTCA---ACCGAATCATTCGGCGTTTCGTGACTAACTGAAA----- | |
| droWil2 | scf2_1100000004382:576178-576223 + | GCCTTTGCGGGTCAGGATCTGTTAGATGATGTATAATGTGATCTTC-------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12875:2678655-2678686 + | GCGATC-CGGTTCCGGATCATCATGATGCATCA--------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15112:3492067-3492158 - | GCGCTC-CGGGTTGGGATTGGTATAGTCATCCAGCACCACATTGG------CGCCATATGCCCAAAGTTGATTATAGGCCAAGAGCCAGAACAGCAGCA--------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 12/06/2013 at 02:35 PM