| droSim3 |
2R:1784908-1785065 + |
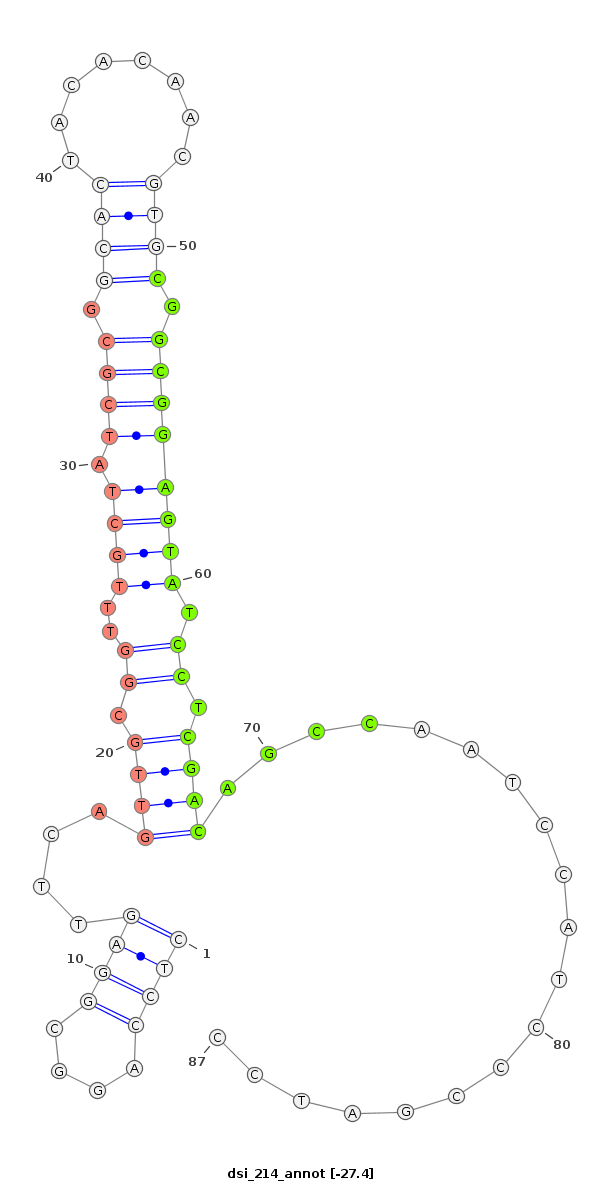
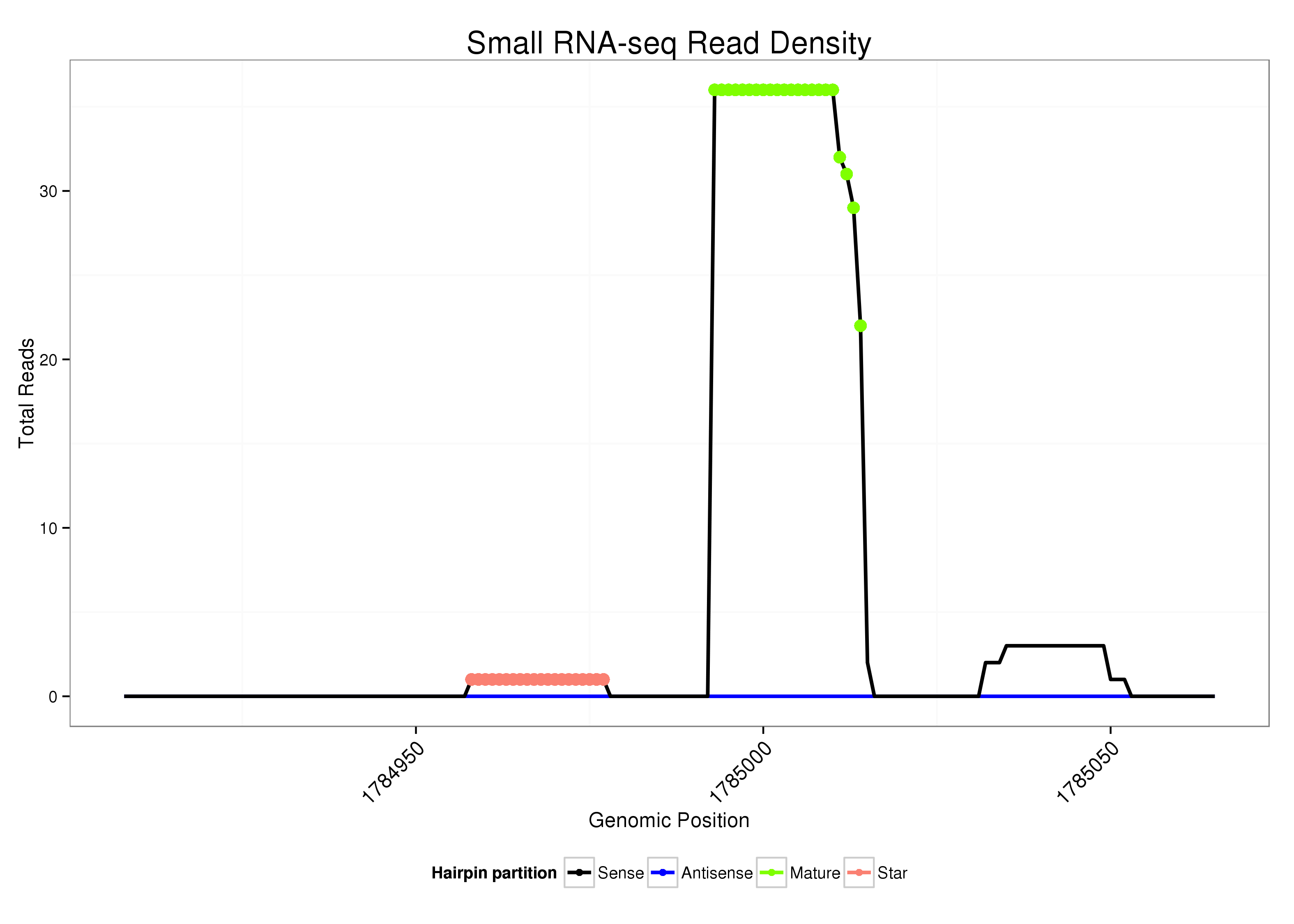
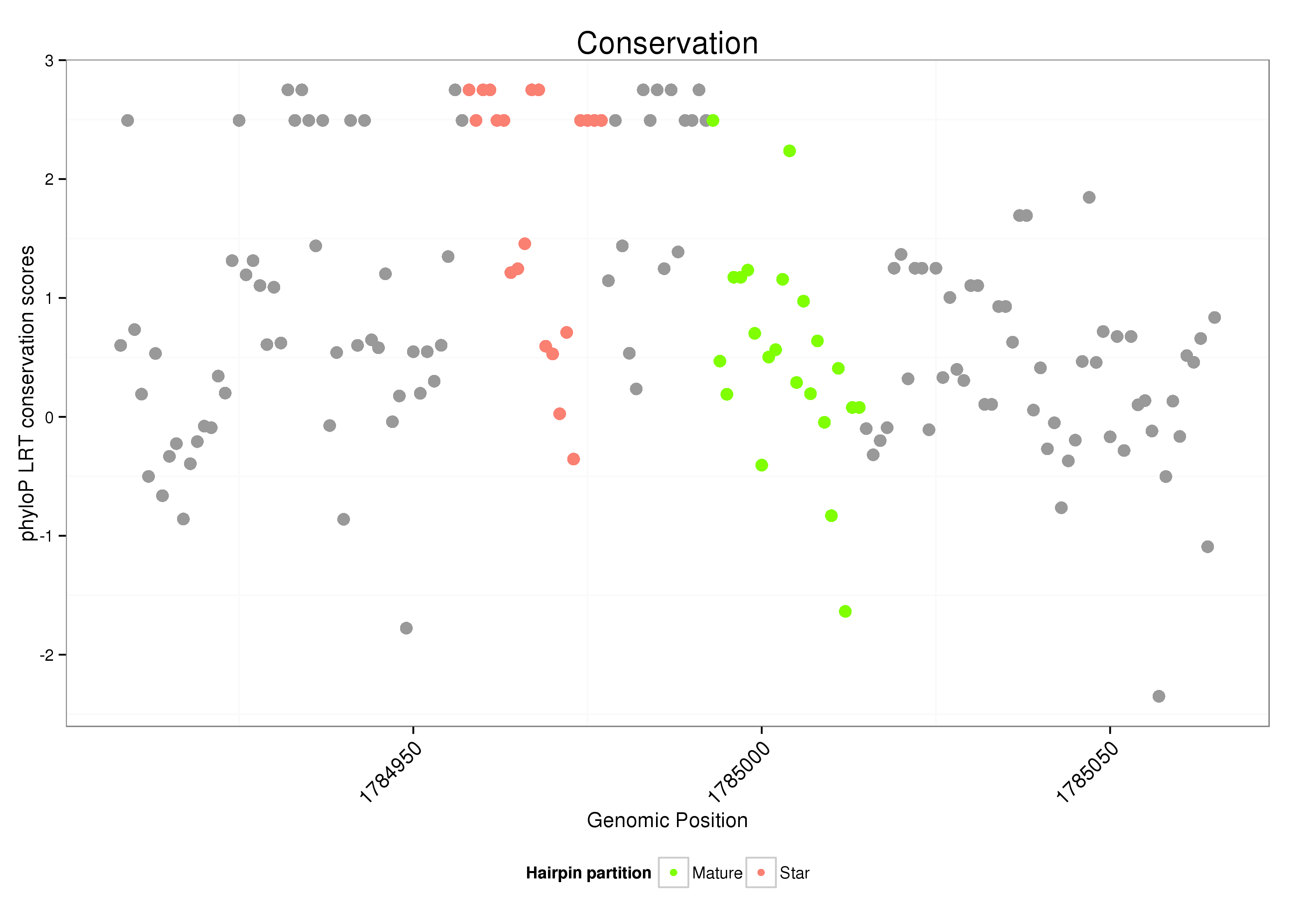
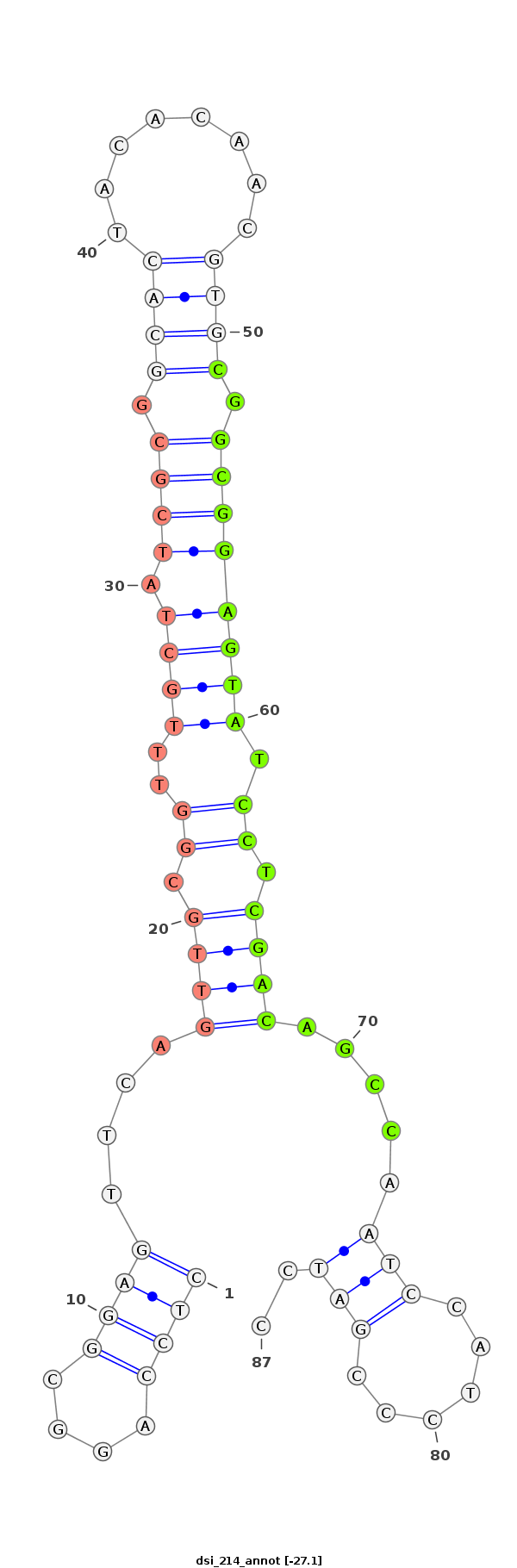
dsi_214 |
CCAGGAC------------GATGGAGTGTCCTGGCCAGACAGCCCGTCTCCA-------------------------------------------------------------------G-GCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGGAGTAT------CCT---------CGACAGCC---------------------------AA--------------------TCCATCCCGATCCTTCCTT---------------TCGCGACTCTCG---------A--ATCGCGACTCT-----TCGCGT |
| droSec2 |
scaffold_1:655265-655415 + |
|
CCAGAAC------------GATGGTGTGTCCTGGCCAGACAGCCCGTCTCCA-------------------------------------------------------------------G-GCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGGAGTAT------CCT---------CGACAGCC---------------------------AA--------------------TCCATCCCGATCCTTCCTT---------------TCGCGACTCGCG---------A---------CTCT-----TCGCGT |
| dm3 |
chr2R:2986013-2986170 + |
|
CCAGGAC------------GATGGAGTGTCCTGCCCAGACAGCCCGTCTCCA-------------------------------------------------------------------G-GCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGGAGTAT------CCT---------CGACAGCC---------------------------AA--------------------TCCTTCCCGAACCTTCCTT---------------TCGCGACTCACG---------A--CTCTCAACTCT-----TCGCGT |
| droEre2 |
scaffold_4929:19580945-19581077 - |
|
CCAGGAC------------GACGGAGTGTCCTGGCCAGACAGCCCGTCTCCA-------------------------------------------------------------------T-GCGG-------CGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGCAGTAT------CCT---------CGACAA--------------------------------------------------------------GCTTCC----------------TGCGCGATTCCCG---------A----------G-C-----TCGCGT |
| droYak3 |
2L:15698324-15698456 + |
|
CCAGGAC------------GATGGAGTGTCCTGACCAGACAGCCCGTCTCCA-------------------------------------------------------------------T-GCGG-------AGTTCAGTTGCGGCTTGCTAACGCGGCACTACACAACGTGC-GGCGGAGTAT------CCT---------CGACAT--------------------------------------------------------------CCTTCC----------------TGCGCGATTTCCG---------A-----------CT-----TCGCGT |
| droEug1 |
scf7180000409672:5008822-5008974 + |
|
CCAAGACGGAAAAC----------AGAGTCCTGGCCAGACAGTCAGTCTCCA-------------------------------------------------------------------G-TCGG-------AGTTCAGTTGCGGTTTGCTAACGCGGCACAACACAACGTGC-GGCGGAGTAT------CCT---------CGATATCCTCGATAT-------------------------ATCCTTT--ACATCCTCGACATCCCGGTCCTTCC----------------TTCGC------------------------------------------ |
| droBia1 |
scf7180000302292:3154014-3154153 - |
|
CCAGGTCCTGAGACAGAGTCGCGGACCATCCTGGCCAGACAGTCGGTCTCCA-------------------------------------------------------------------G-CCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGGAGTAT------CTT---------TGAGCTCCCAGAT---------------------------A-----------T---------------------------------------------TCTCG---------A----------A-T-----ATCCTC |
| droTak1 |
scf7180000415382:228079-228239 + |
|
CCAGAAGGGAAAACCGAGCCACGGAGTATCCTGGCCAGACAGTCAGTCTCCG-------------------------------------------------------------------G-CCGG-------AGTTCAGTTGCGGTTTGGTATCGCGGCACTACACAACGTGC-GCCGGAGTATCAGTATCCT---------CGAAAACC---------------------------TT---------G--ACATCCTCGACATCCCGATCCTTCC----------------ATCGCGTC--------------------------------------- |
| droEle1 |
scf7180000491265:108274-108483 + |
|
CCAGGAGCGGAAGCAGCGCCACGGAGTGTCCTGCCCAGACAGCCAGTCTACG-------------------------------------------------------------------G-GCCG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC--GCGGAGTGT------CCT---------CGGTATCCTCAGTAT----CCTCGGTATCCTCGAC-----ATCCTCG--ACATCCTCGACAGCCCGATCCTTCT-----------GTTCCTTCGCGTTGTTCG---------ACTTTCTCGA-TTG-----TCGCGT |
| droRho1 |
scf7180000780072:163374-163551 - |
|
CCAGGACCGAAAACAGAGCCACGGAGTGTCCTGGCCAGACAGTCAGTCTCCG-------------------------------------------------------------------G-CCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACATAACGTGC--GCGGAGTAT------CCT---------CGAAATCCTCGAC---------------------------ATCCTCG--ATATCCTCGACATCCAGATCCTTTC----------------TTCGCGTTCCTCG---------A----------CTT-----TCGCGT |
| droFic1 |
scf7180000453955:273999-274167 - |
|
CCAAGACAGAAAGCAGAGCCACCAAGTATCCTGGCCAGACAGTCAGTCTCCG-------------------------------------------------------------------G-CCGG-------AGTTCAGTTGCGGTTTGCTATCGCGGCACTACACAACGTGC-GGCGGAGTAT------CCT---------CGAAGTCC---------------------------TA---------G--ATATCCTCGACATCCCGATCCTTCC-----------------TCGCGTTCCTCG---------A----------GTT-----TCGCTT |
| droKik1 |
scf7180000302640:92221-92370 + |
|
CCAGGAC------------GCGCAAACATCCT-GCCAGACTGCCGGGCCCCAG----------------------------T-------------------------------------G-GCGG-------AGTTCAGTTGCTATTTGCCAGCGCGGCTCTACACAACGTGC-GGCGGACTTG--CTATCCTCT-------CGACAA--------------------------------------------------------------CG----TTCA-------GTTCCATCGAGCTCCCCGAAT--------------TA-CGT-----ACGCGT |
| droAna3 |
scaffold_13266:3223073-3223233 + |
|
GCAGAAT----------------CAGTGTCCTGGCTAGACAGCCCGGCTCCG-----------------------------T-------------------------------------G-ACAG-------CGTTCAGTTGCGGTTTGCTTACGCGGCAGAACACACCGTGC-GGCGGCCTAT------CCT---------CGCCATCC---------------------------TT--------------------TGCATCCTGA------------------GATCCTTCGTCCTTTTGGCATTTTATTT--TACGCGTTCCT-----TCGGCT |
| droBip1 |
scf7180000396427:1694036-1694201 + |
|
TCAGCAG-------------AAGCGTTGTCCTGGCTAGACAGACCGGCTCCG-----------------------------C-------------------------------------G-ACAG-------CGTTCAGTTGCGGTTTGCTAACGCGGCACAACACACCGTGC-AACGGCTAAT------CCT---------TGCCAATT---------------------------TT--------------------GGCATCCTGA------------------GATCCTTCGTCTATTTGGCATTT---TT--TACGCGTTCCTCCGGCTCGCTT |
| dp5 |
3:1271504-1271677 + |
|
CCACCA-----AGCAGCGT-GCAGCGTGTCCTTGCCAGACAGCCGGGCTCCG-------------------------------------------------------------------A-ACGGAGTCCGTAGTTCAGTTGCGGTTTACAAGCGCGCCACTACACAACGTGCAGGCGGAGTATAGCTATCTTTATCCCAGA---CA-----------------------------------------------T---------------CC----TTTGGCACGCGCTTCCCTCGTGTTCC--GTTT--------------GA-TCT-----ACGCGT |
| droPer2 |
scaffold_2:1446966-1447139 + |
|
CCACCA-----AGCAGCGT-GCAGCGTGTCCTTGCCAGACAGCCGGGCTCCG-------------------------------------------------------------------A-ACGGAGTCCGTAGTTCAGTTGCGGTTTACAAGCGCGCCACTACACAACGTGCAGGCGGAGTATAGCTATCCTTATCCCAGA---CA-----------------------------------------------T---------------CC----TTTGGCACGCGCTTCCCTCGTGTTCC--GTTT--------------GA-TCT-----ACGCGT |
| droWil2 |
scf2_1100000004513:2577721-2577936 + |
|
CCGG-------------------TAGTGTCCTTGGTAGACAGCACGGCTCCAGCTCCAGCTCCGGCGCACAGTTCCAACTCCGACAAAAAACTTTGACTCCGGTTTGGTTTCAGTTTCAGAATGAGTTCA--GTTTCAGTTGCGGTTTGCTAGCGCGGCAAAACACAACGTGCAGGCGGAAAAT------CCA---------CTTCAATTTCTACT-TGGGCAACG-----------AATATATTCGCTACGCA---------------------------------------------TATTCG---------A---------------------CTT |
| droVir3 |
scaffold_10324:1142317-1142425 + |
|
CCAGGCA----------------TGTTGCCACTGGCAGACAGCGTGTCCTTGTCTCCGGCTTCTGGCA---------------------------------------------------G-CGGC-------AAATCAGTTGCGGTTTTCAGACGCGGCACAACACAACGTGC-AAGCGGATAT------C---------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:4898037-4898141 - |
|
CCGGGCA----------------TGTTGCCACTGGCAGACAGCGTGTCCTTGTTTCTGGCTTTTGGCCA--------------------------------------------------T--GGC-------AAATCAGTTGCGGTTTGGTAACGCGGCACAACACAACGTGCAAGCGGA--------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15245:9224393-9224497 + |
|
CCAGGCA----------------TGTTGCCACTGGCAGACAGCGTGTCCTTGTCTCCGGCTTCTGGCT---------------------------------------------------G-TGTC-------AAATCAGTTGCGGTTTGCAAACGCGGCACAACACAACGTGCAAGCGGA--------------------------------------------------------------------------------------------------------------------------------------------------------------- |