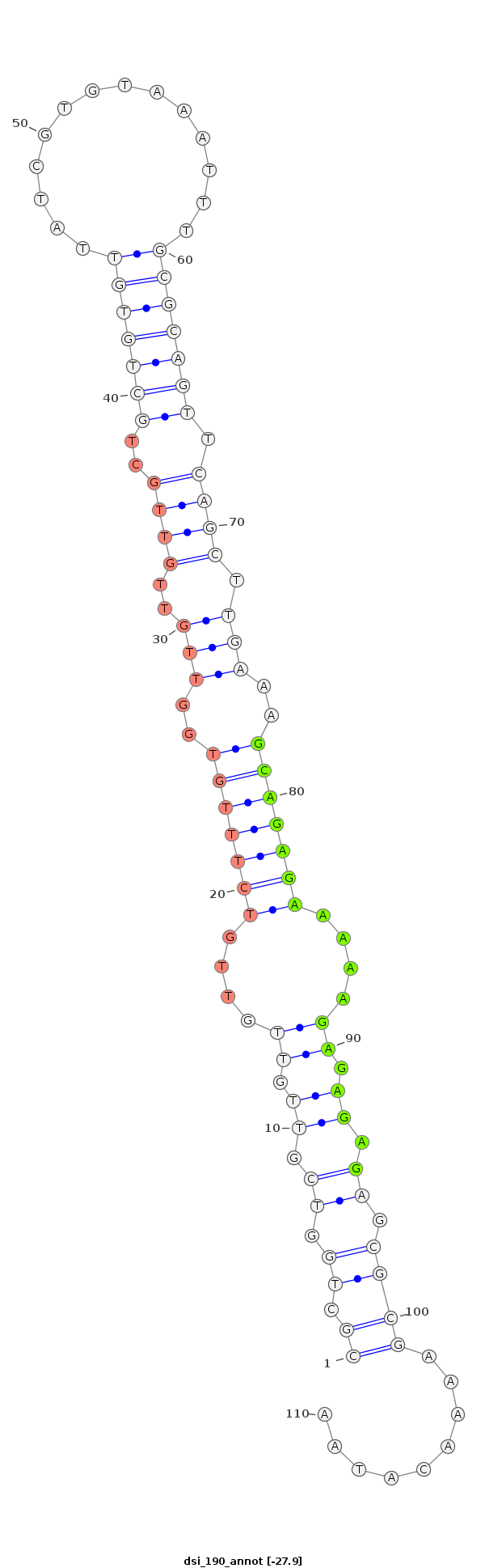
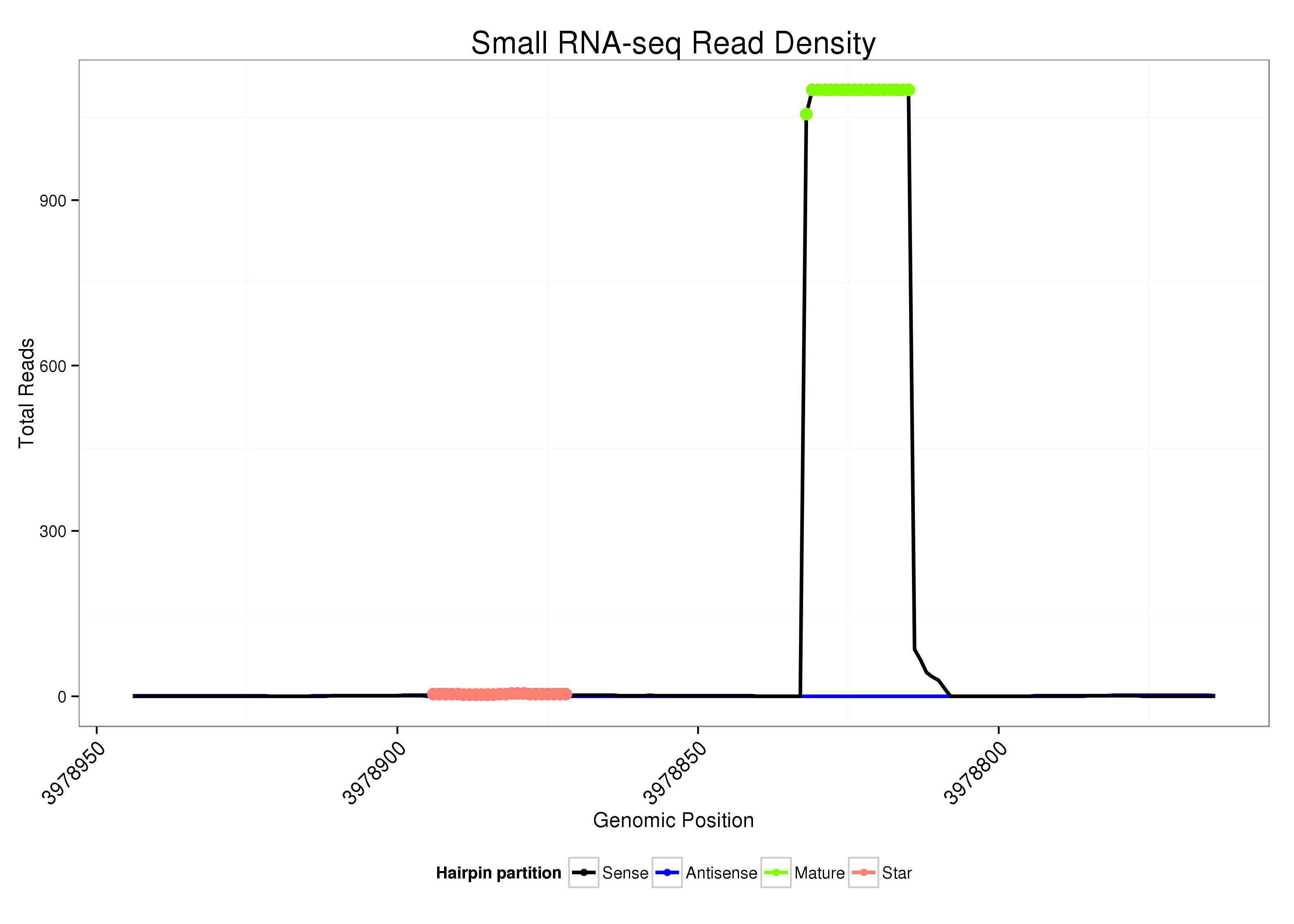
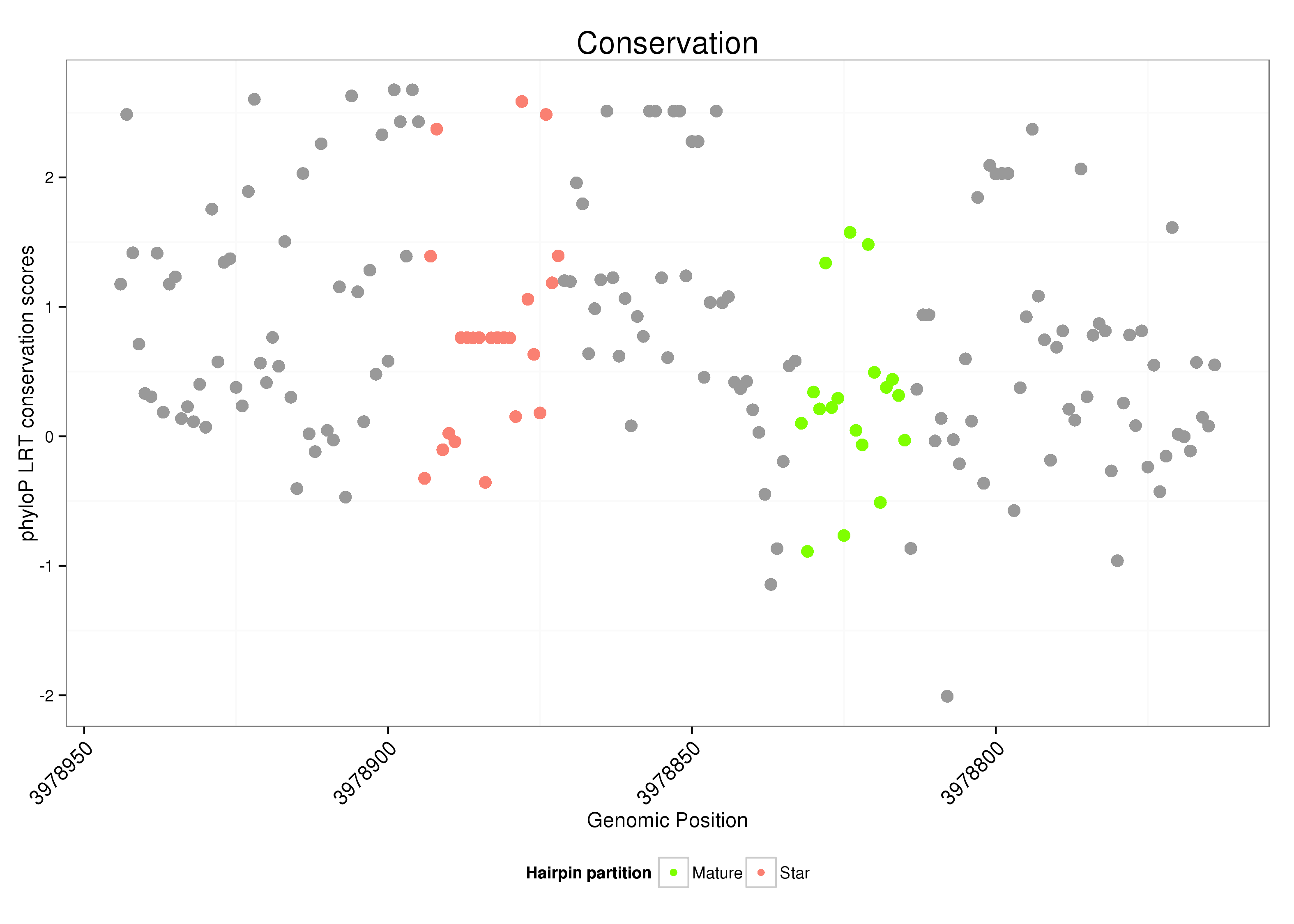
ID:dsi_190 |
Coordinate:2R:3978814-3978894 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -27.5 | -27.4 | -27.2 |
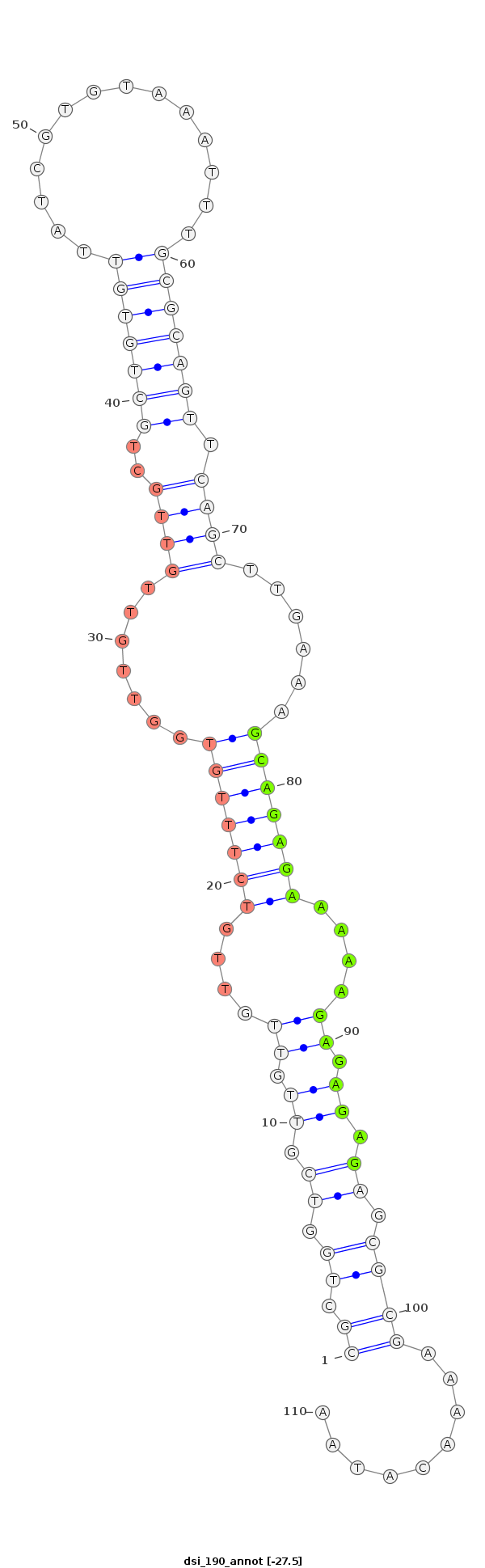 |
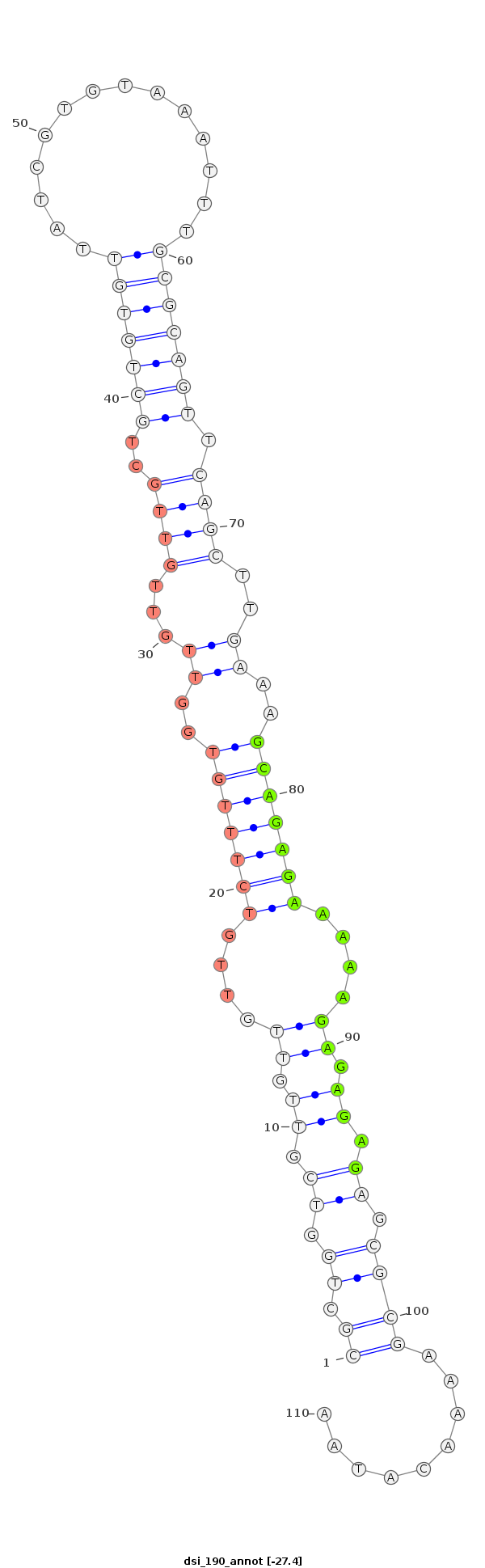 |
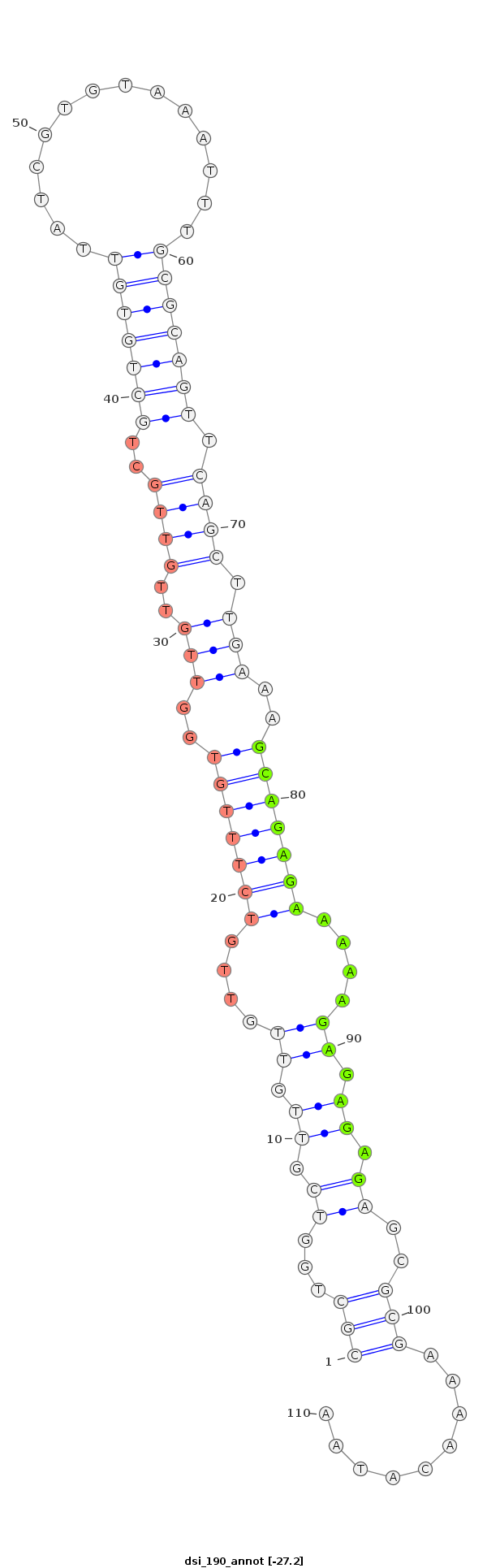 |
intergenic
No Repeatable elements found
|
GGATATTCCACAGAGCACACTGTTGTTGTTGTTGTCGCTGGTCGTTGTTGTTGTCTTTGTGGTTGTTGTTGCTGCTGTGTTATCGTGTAAATTTGCGCAGTTCAGCTTGAAAGCAGAGAAAAAGAGAGAGAGCGCGAAAACATAAAACCAAACCGTTGTCTTTTAAACTCTCCCCGCCAGC
***********************************((.((.((.((.((....(((((((..(((..((((..(((((((..............))))))).)))).)))..)))))))....)).)).)).)))).........************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
O001 Testis |
M023 head |
SRR553485 Ovary |
GSM343915 embryo |
SRR553487 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................GCAGAGAAAAAGAGAGAG................................................... | 18 | 0 | 2 | 1015.00 | 2030 | 1814 | 216 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGA.................................................. | 19 | 0 | 2 | 17.00 | 34 | 28 | 6 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGAGCGC.............................................. | 22 | 0 | 1 | 14.00 | 14 | 14 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGAG................................................. | 20 | 0 | 2 | 13.00 | 26 | 24 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGAGCGCG............................................. | 23 | 0 | 1 | 11.00 | 11 | 10 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGAG................................................. | 19 | 0 | 2 | 10.00 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGAGC................................................ | 21 | 0 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGAGC................................................ | 20 | 0 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGAGCGCG............................................. | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGAGCG............................................... | 22 | 0 | 2 | 3.00 | 6 | 4 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGAGCG............................................... | 21 | 0 | 2 | 3.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TTGTCTTTGTGGTTGTTGTTGCT............................................................................................................ | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGAGAAAAAGAGAGAGA.................................................. | 18 | 0 | 3 | 2.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TGTTGTTGCTGCTGTGTTATCGTG.............................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GTAAATTTGCGCAGTTCA............................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................GTTGTTGTTGCTGCTGTGTT.................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................GCAGAGAAAAAGAGAGAGAGCGC.............................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................GTCGCTGGTCGTTGTTGTTGTC.............................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................TGTTGTTGTCTTTGTGGTTGT................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AACCGTTGTCTTTTAAAC............. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................AATACATTAAACTAAACCGTTGT...................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................AGCTTGAAAGCAGGGA.............................................................. | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................GCTGGTCGTTGTTGTTGG............................................................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................AACAGTTGTCTTTTAA............... | 16 | 1 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ...........ATACCACCCTGTTGTTGTT....................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGTGGTTGTTGTTGCTGCT......................................................................................................... | 19 | 0 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................TGTTGTCTTTCTGGTAGTGGT................................................................................................................ | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GCTGGCGGGGAGAGTTTAAAAGACAACGGTTTGGTTTTATGTTTTCGCGCTCTCTCTCTTTTTCTCTGCTTTCAAGCTGAACTGCGCAAATTTACACGATAACACAGCAGCAACAACAACCACAAAGACAACAACAACGACCAGCGACAACAACAACAACAGTGTGCTCTGTGGAATATCC
************************************.........((((.((.((.((....(((((((..(((.((((.(((((((..............)))))))..))))..)))..)))))))....)).)).)).)).))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O002 Head |
SRR553485 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CAGTGTGCTCTGTGGAATATC. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ..............................TTGGTTTTATGTTTTCGCG.................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................GTGCTCTGTGGAATATCC | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| GCTGGCGGGGAGAGTTTAAAAGA.............................................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ..................................................TCACTCTCTTTCTCTCTGCTAT............................................................................................................. | 22 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 |
| ........GGAGAGTTGAAAAGA.............................................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| .................................................................................................................................AACAGGAACGACCAGCG................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .................AAAAGACCTCGGTTTGGT.................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................ACAGGAACGACCAGCG................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ....................................................TCACTCTTTTTCTCTCCT............................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 2R:3978764-3978944 - | dsi_190 | GGATATTCCACAGA-GCACACTGTTGTTGT-------------T---GTTGTCGCTGGTCGTTGTTGTTGTCTTTGTGGTTGT------TGTTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGAAA----------------------------------------------G-C-AGAGAAA---------------------------------AA-GAGAGA-------------------------------------------------GAGCGCGAAAACAT-AAA--------ACCAAACCGTTGTCTTT---T--------------------AAACTCTC--------------CCCGCCAGC |
| droSec2 | scaffold_1:2953947-2954139 - | GGATATTCCACAGA-GCACACTGTTGTTGT----TGTTGTTGTTGTTGTTGTCGCTGGTCGTTGTTGTTGTCTTTGTGGTTGT------TCTTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGAAA----------------------------------------------G-C-AGAGAAA---------------------------------AA-GAGAGA-------------------------------------------------GAGCGCGTAAACAT-AAA--------ACCAAACCGTTGTCTTT---T--------------------AAACTCTA--------------CCCGCCAGC | |
| dm3 | chr2R:5328847-5329004 - | GGATATTCCACAGA-GCACACTGTTGTTGT-------------T---GTTGTCGCTGGTCGTTGTTGTTGTCGTTGTGGTTGT------TGTTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGAAA----------------------------------------------G-C-AGAGAAA---------------------------------AA-GAGAGA-------------------------------------------------GAGCGCGAAAAAAT-AAAA-------A-----------------------------------------------A--------------CCCGCTAGC | |
| droEre2 | scaffold_4929:9022156-9022334 + | GGATATTCCATATA-GCACACTGTTGTTGT-------------------TGTCGCTGGTCGTTGTTGTTG---------------------TTGCTGCTGT--GTTACC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGAAA----------------------------------------------G-C-TGAGAAA---------------------------------AA-GAGAGA--------GAG-------A--G----AGAG----------------A--GCGCGCGAAAACAT-AAAAATAA--AACTAAACCGTTGTCTTT---T--------------------AAACTCTC--------------CCCGCCAGC | |
| droYak3 | 2L:17978055-17978227 - | GGATATTCCACATA-GCACACTGTTGTTGT-------------------TGTCGCTGGTCGTTGCTGTCG---------------------TTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGAAA----------------------------------------------G-C-TGAGAAA---------------------------------AA-GA--------AGGAGAT-------A--A----AGAG----------------A--GAGAGCGAAAACAT-AAA--------ACTAAACCGTTGTCTTT---T--------------------AAACTCTC--------------CCCGCCAGC | |
| droEug1 | scf7180000409183:959219-959405 - | GGATATTCCACATA-GCACACTGTCGTTGT-------------TGTTGTCGTTGCTGGTCGTTGTTGCTG------------T------TGCTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCCTAAAA-----------------------------------------------AG-AGAGATA---------------------------------GA-GG--------AAGAGCC-------T--AAA--AGAG----------------AGCTCACGCGCAAAAATCAAA--------ACTAAACCGTTGTTTCT---T--------------------TGACTCTC--------------CCTGTCAGC | |
| droBia1 | scf7180000301506:1389643-1389829 + | GGATACTCCACATA-GCACACTGTTGTTGT-------------------TGTCGCTGGTCGTTGTTGTTG------------T------TGCTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTTGGAA-----------------------------------------------AG-AG--T------------------------------------------GTGAGAGCGAGATAGAAACCCAGAAA--AGAG----------------AGCTCGCGCGCTAAAACAAAA--------ACT-AACCGTTGTTTTT---G--------------------TAACTCTC--------------TCTGCCAGC | |
| droTak1 | scf7180000415401:370273-370449 - | GGACTTTCCACATA-GCACACTGTTGTTGT-------------------TGTCGCTGGTCGTTGTTGTTG------------T------TGCTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCTGGAAA-----------------------------------------------AG-AGAGAGA---------------------------------AA-GA------------C---------CAGAGA--AGAG----------------AGCTCGCGCGCTGAAACAAAA--------ACTAAACCGTTGTTTTT---G--------------------TAACTCTC--------------CCTGCCAGC | |
| droEle1 | scf7180000491107:1783408-1783561 + | GGATATTCCCCATA-GCA--CTGTTGTTGT----------TGTAGTTGCTGTCGCTGGCCGTTGTTGTT-------------T------TGCTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGCCTGGAA-----------------------------------------------AG-AG--------------------------------------------AGA-------------------------------------------------GAGCGG----------------------AAAATCGTTGTTTTT---G--------------------AAAATCTCCCA---------ACCCTACCAGC | |
| droRho1 | scf7180000777217:217611-217754 - | GGATTTTCCACATA-GCAC--TGTT----------------------GTTGTCGCTGGTCGTTGTTGTTG------------T------TGCTGCTGCTGT--GTTATC---------G----------TGT---AAA-TTTGCGCAGTTCAGATTGAAA-----------------------------------------------AGAAG--------------------------------------------AGA-------------------------------------------------GAGCA----------------------GAAAACCGTTGTATTT---G--------------------TAACTCTCCCT---------TCCCTACCAGC | |
| droFic1 | scf7180000453858:1365244-1365379 + | GGATATTTCACATA-GCACACCGTTGTTGT--------------------------------TGTTGCTG------------C------TGCTGCTGTTGT--GTTATC---------T----------TGT---AAT-TTTGCGCAGTTCAGCTAAAAA-----------------------------------------------AG-AG--AGA---------------------------------A----------------------------------AGAG----------------AGCTCGCGCGAT--------A--------AGTAAACCGTTGTTTTT---G--------------------TAACT----------------------CAGC | |
| droKik1 | scf7180000302476:707483-707567 - | GGATATTACACTCG-AC------------------------------------AGTGGTTGGTGTTGTTG------------T------TGCTGCTGCTGG--CTTAAC---------G------------T---AAT-TTCGCAAAG----------AG-------------------------------------------------------------------------------------------G-GACTGA-------------------------------------------------GAGCGCCGAG---------------------------------------------------------------------------------------- | |
| droAna3 | scaffold_13266:293453-293642 + | GGATACTCCATTCGACCGCATTGTTTCA--TG-CTGGTGCTGCTGCTGCTGTTGTTGG------------------------A------TGTAGCTGCTGGATGTTTTT---TTTATTTTTAAATAAAAATTAAAATA-TTTGCACAAAATAGTGCCTAA----------------------------------------------G-------T-------------------------------------------------------------------------GTGTGAGTGAAAAGGAAA--GAGCGTGATAA----A-------------AAACCCCCCTTTTC---G--------------------G--CCATCCCA---------GCCCCACCTAG | |
| droBip1 | scf7180000396730:1316831-1317032 + | GGATACTCCATTCGACCGCATTGTTTCTGC-------------T---GCTGCTGCTGTTGGATGTTGCTG---------------------TTGGAT------GTTTTT-----TATTTC-------GATTTGAAATA-TTTGCGCAGTTTGCCAAAATA---GT-----------------------------------------G-C-CTAGT-------------------------------------------------------------------------GCGTGAGTGAGAAGGCAA--GA-----CTGTAATAAAA--------ACTTATCGGTTGTTTTTTTTTAAAGCCGAGAAACGTCCGCAAAACTGCT--------------GCCGTTGCC | |
| dp5 | 3:13989368-13989539 + | GGATATTCCACATC-GCA--TGGTTGTTGT-------------A---GTAGA-GGTGGTTGTTGTTGCTG------------TTGCTGCTGCGGCTGC--T--GTTATC---------G------------T---AAATTTTGCGCAGTTCG-TTCGCTA----------------------------------------------A-T-AGAAAAA---------------------------------AATGAGAG-------GAGAGGGA----CAGCGACTAGAG----------------A--GCGCGCGCA------AAA--------ACTAAACTGTATTTTTT---T--------------------GAATT-------------------------- | |
| droWil2 | scf2_1100000004510:3178112-3178178 - | TGGGGCAAA------------TGTTGT----------------AGATGTTGACGCTGGTTGTTGTTGTTGTTGTTGTTGTTGT------TGCTGCTGCTGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_10324:87657-87773 - | GGATATTCCAT-CG---------TCA---GT----------------GTCGATG-TGGTTGTTGTTGCTG------------C------TGCGGCTGC--T--GCTATCTTGTTTG--T----------TGT---AAA-TTTGCGCAGTACATTTCAAAT-----------------------------------------------AA-AA--A------------------------------------------TAAAGCACGAAAC-------------------------------------------------AAA-AA-------------AA------------------------------------------------------------------ | |
| droMoj3 | scaffold_6496:1908056-1908186 - | GGATATTCCAT-CG---------TCA---GTGT----------C---GATGT-GTTTTTTGTTGTTGCTG------------C---TGCTGCGGCTGC--T--GTTATCTTGCTTG--T----------TGT---AAA-TTTGCGCAGTAAATTAAGAAA-----------------------------------------------AT-A----AA---------------------------------AC-GAAAT----------------------------------------------------ACGACAAAAAAT-AAA--------AATAAA------------------------------------------------------------------ | |
| droGri2 | scaffold_15245:15540354-15540616 + | GGATATTCCAT-CG---------TCA---GT----------------GTCGATG-TGATTGTTGTTGTTGCTG---------C------TGCGGCTGC--T--GTTATCTTGCTTG--T----------TGT---AAA-TTTGCGCAGTACATTTAAAAAAAAACGCCACACAAACAAACCAAAAAAAAAGTCCGCATAATACAGAAAG-AG--AG-CGATAGAGAGCCAGACAGGAGGAGATAGAGATAG----------------------------------AGAG----------------AGCGCGCGCGAAAAGAA-AAAAATAATAAAGTAAACGGTTGTTGTTTTTG--------------------TGTTTTTGTATTTTGTACCAG--CCG-CAGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 12/06/2013 at 02:34 PM