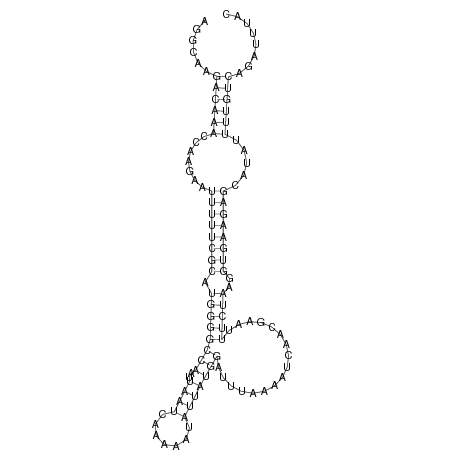
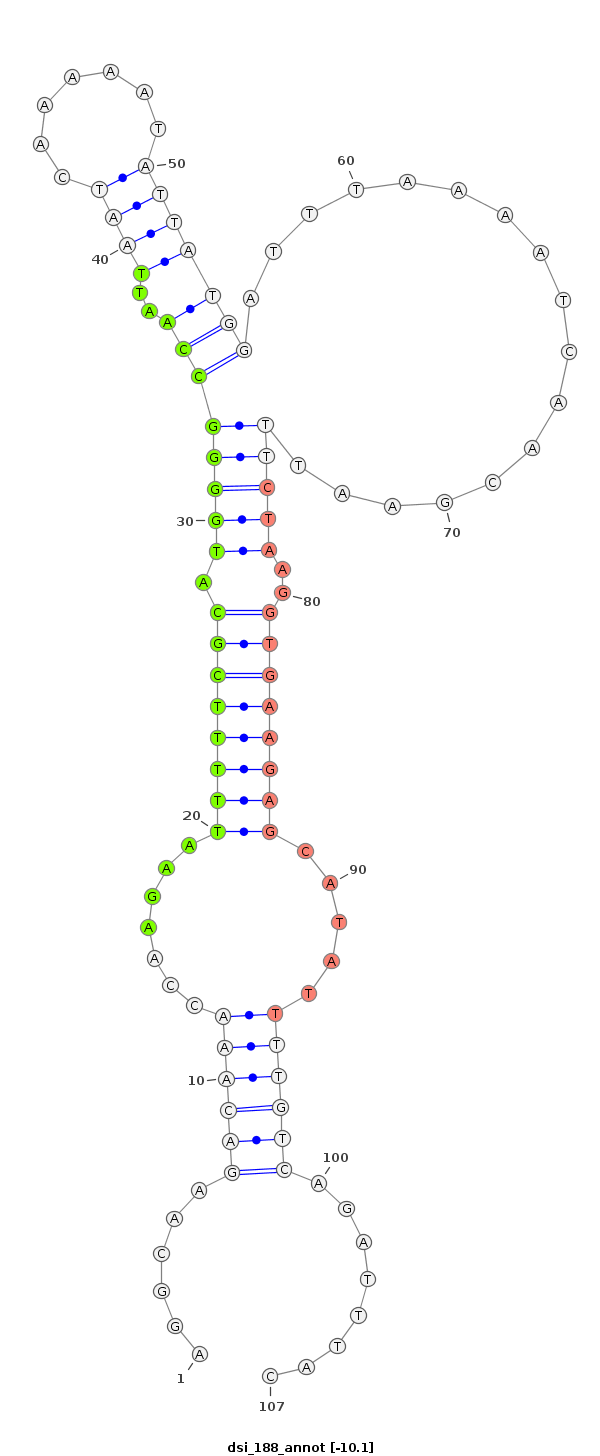


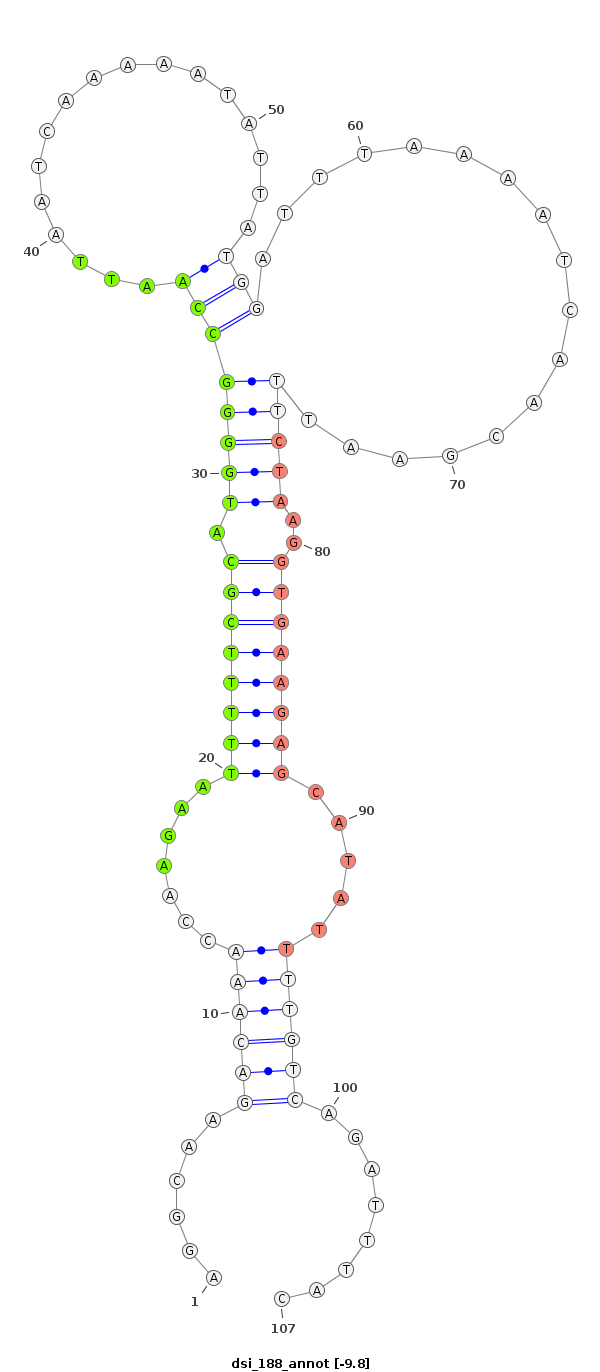
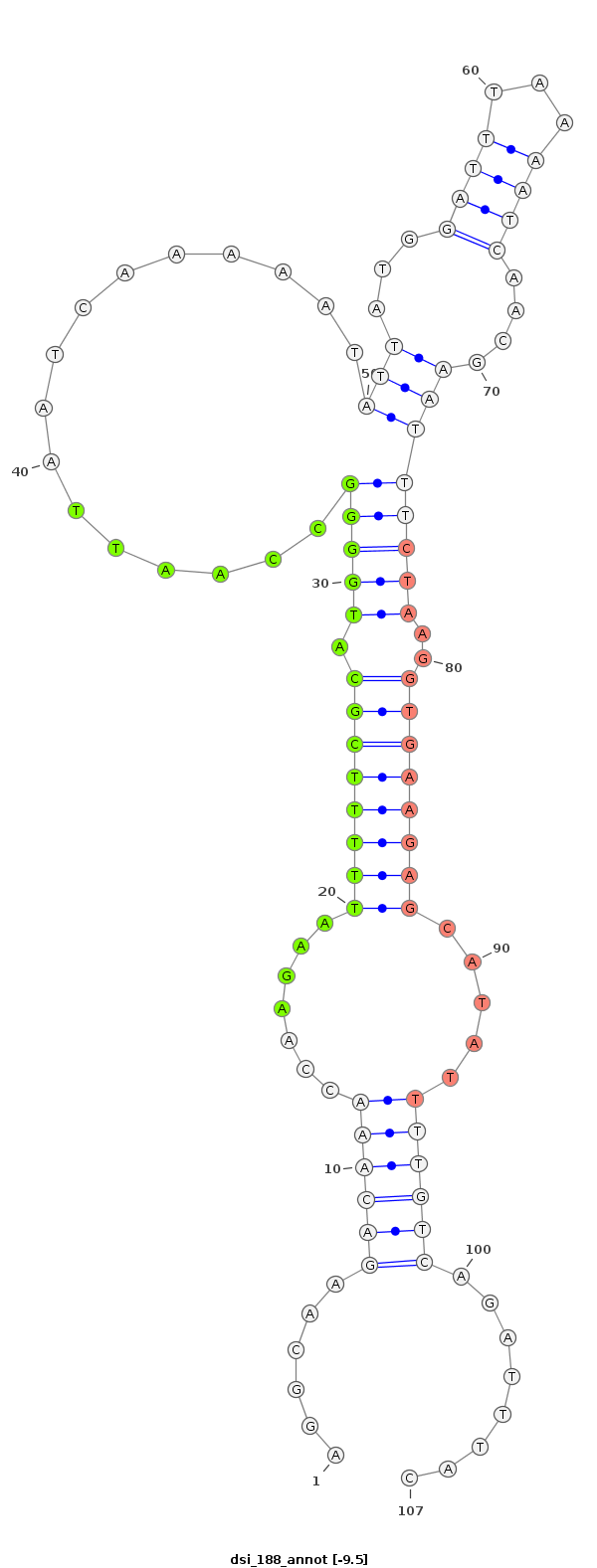
ID:dsi_188 |
Coordinate:chrU_M_105:896-982 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -9.8 | -9.5 | -9.4 |
 |
 |
 |
intergenic
No Repeatable elements found
|
AGGCAAGACAAACCAAGAATTTTTCGCATGGGGCCAATTAATCAAAAATATTATGGATTTAAAATCAACGAATTTCTAAGGTGAAGAGCATATTTTGTCAGATTTACAAGGCATGAGCACACGTATGCACACATACAGTTGTA
******************************************......((((((.......((((((((.((((((((..((((.......))))))).................)))))..)))))))).....))))))........************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR553485 Ovary |
SRR618934 Ovary |
SRR553487 Ovary |
SRR553486 Ovary |
M024 male body |
M025 embryo |
M053 Female-body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................... | 18 | 0 | 20 | 17.60 | 352 | 352 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGCCAA.............................................................. | 24 | 0 | 1 | 9.00 | 9 | 8 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGG................................................................... | 19 | 0 | 2 | 8.00 | 16 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTCTAAGGTGAAGAGCATA....... | 19 | 0 | 2 | 4.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGG.................................................................. | 20 | 0 | 2 | 2.50 | 5 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................AAGGTGAAGAGCATATTTTGTC | 22 | 0 | 3 | 2.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGCC................................................................ | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGCCA............................................................... | 23 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 17 | 0 | 20 | 1.85 | 37 | 0 | 0 | 37 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGCCAATTA........................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTCTAAGGTGAAGAGCATATT..... | 21 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGCCAAT............................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AACGAATTTCTAAGGTGA............... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................CTAAGGTGAAGAGCATAT...... | 18 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGAATTTCTAAGGTGA............... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGGCAATT............................................................ | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGC................................................................. | 21 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 30 | 0 | 20 | 0.70 | 14 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTCTAAGGTGAAGAGCA......... | 17 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATAGGGCCAAT............................................................. | 25 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 20 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................ATTTTGGATTTAAAATCATC.............................. | 20 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATG..................................................................... | 17 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ........................................................ACAAGAATTTTTCGCATGGGGCCAA.............................................................. | 25 | 1 | 20 | 0.15 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................TTAAAGTCAACGAATTTCTAAGGTG................ | 25 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................................................................TCTAAGATGAAGAGCAAA....... | 18 | 2 | 16 | 0.13 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTCTAAGATGAAGAGCAAATTT.... | 22 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TCTAAGATGAAGAGCAAATT..... | 20 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 29 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ............................................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .........................................................CAAGAATTTTTCGCATGGGGTCAAT............................................................. | 25 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................ACAAGAATTTTTCGCATGGGGCCAAT............................................................. | 26 | 1 | 20 | 0.10 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AACGAATTTCTAAGGTGAAGGGC.......... | 23 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................CTAAGATGAAGAGCAAAT...... | 18 | 2 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TAAGATGAAGAGCAAATTT.... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TAAAGTTAACGAATTTCTAAGGTGA............... | 25 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGTCAACGAATTTCTAAGG.................. | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................... | 30 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................... | 15 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 16 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TAAAGTCAACGAATTTCTAAGGTGAA.............. | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................... | 30 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TAAAGTCAACGAATTTCTAATGTG................ | 24 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................TAAGTCAACGAATTTCTAAGGTG................ | 23 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TACAACTGTATGTGTGCATACGTGTGCTCATGCCTTGTAAATCTGACAAAATATGCTCTTCACCTTAGAAATTCGTTGATTTTAAATCCATAATATTTTTGATTAATTGGCCCCATGCGAAAAATTCTTGGTTTGTCTTGCCT
************************************........((((((.....((((((((..(((((.................(((((((.......))))..)))))))).)))))))).......))))))......****************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 Ovary |
M053 Female-body |
O002 Head |
SRR553487 Ovary |
SRR553485 Ovary |
SRR553488 Ovary |
SRR553486 Ovary |
GSM343915 embryo |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................TGACAAAATTTGCTCTTCACCTTAGA.......................................................................... | 26 | 1 | 2 | 16.50 | 33 | 0 | 0 | 0 | 7 | 22 | 3 | 1 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGTGTGC.................................................................................................................... | 25 | 0 | 20 | 3.20 | 64 | 19 | 5 | 32 | 0 | 6 | 0 | 0 | 2 | 0 |
| ..CAACTGTATGTGTGCATACGTGTGCTCA................................................................................................................. | 28 | 0 | 20 | 1.65 | 33 | 2 | 12 | 0 | 2 | 5 | 8 | 4 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTCATGC.............................................................................................................. | 27 | 0 | 20 | 1.55 | 31 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATTTGCTCTTCACCTTA............................................................................ | 24 | 1 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCTCTTCACCTTAGA.......................................................................... | 26 | 0 | 2 | 1.00 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........TGTGTGCATACGTGTGCTCATG............................................................................................................... | 22 | 0 | 20 | 1.00 | 20 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGTGTGCTCA................................................................................................................. | 27 | 0 | 20 | 1.00 | 20 | 13 | 3 | 0 | 1 | 3 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTCA................................................................................................................. | 24 | 0 | 20 | 0.90 | 18 | 0 | 11 | 0 | 2 | 2 | 3 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGT........................................................................................................................ | 20 | 0 | 20 | 0.60 | 12 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCTCTTCACCTTAG........................................................................... | 25 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATTTGCTCTTCACCTTAG........................................................................... | 25 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......GTATGTGTGCATACGTGTGC.................................................................................................................... | 20 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTC.................................................................................................................. | 23 | 0 | 20 | 0.40 | 8 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........TGTGTGCATACGTGTGC.................................................................................................................... | 17 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACTGTATGTGTGCATACGT........................................................................................................................ | 19 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCTTTTCACCTTAGA.......................................................................... | 26 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................TTAGAAATTCGTTGACTTTAAATT....................................................... | 24 | 2 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| .........ATGTGTGCATACGTGTGC.................................................................................................................... | 18 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCTCTTCAACTTAGA.......................................................................... | 26 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCCCTTCACCTTAGAAAT....................................................................... | 29 | 1 | 20 | 0.20 | 4 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTCATG............................................................................................................... | 26 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACG......................................................................................................................... | 20 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTCAT................................................................................................................ | 25 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGTGTGCATACGTGTGCTC.................................................................................................................. | 19 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGT........................................................................................................................ | 21 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| .............GTGCATACGTGTGCTCATGC.............................................................................................................. | 20 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TATGTGTGCATACGTGTGC.................................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............GCATACGTGTGCTCA................................................................................................................. | 15 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............GCATACGTGTGCTCATG............................................................................................................... | 17 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGTGTGCATACGTGTG..................................................................................................................... | 16 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACG......................................................................................................................... | 16 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGT...................................................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGTGTGCTC.................................................................................................................. | 26 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGTGTGCATACGTGTGCTCATGC.............................................................................................................. | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGACAAAATATGCCCTTCACCTTAGAAA........................................................................ | 28 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCT................................................................................................................... | 22 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTAGAAATTCGTTGACTTTAAAT........................................................ | 23 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .............GTGCATACGTGTGCT................................................................................................................... | 15 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTG....................................................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TATGTGTGCATACGTGTGCT................................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGTGTG..................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGTGTGC.................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......GTATGTGTGCATACGTGTGCT................................................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TACAACTGTATGTGTGCATACGTGTGCT................................................................................................................... | 28 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGTGTGCT................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TTACAAAATTTGCTCTTCACCTTAGA.......................................................................... | 26 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......TGTCTGTGTGCATACGTGTGCTCA................................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTCAA................................................................................................................ | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACTTGTGC.................................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TTAGAAATTCGTTGACTTT............................................................ | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....ACTGTATGTGTGCATACG......................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGTGT...................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................TACGTGTGCTCATGC.............................................................................................................. | 15 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GTGTGCATACGTGTGCTC.................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATAC.......................................................................................................................... | 15 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGCGTGCATACGTGTGC.................................................................................................................... | 17 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TATTTTTGATTAATTGG................................. | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGTATACGAGCGC.................................................................................................................... | 25 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............TGCATACGTGTGCTC.................................................................................................................. | 15 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGT........................................................................................................................ | 17 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TTCACCTTAGAAATTCGTTGACTTT............................................................ | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........TGTGTGCATACGTGTGCTCAT................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CTGACAAAATATGCCCTTCACCTTAGA.......................................................................... | 27 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....ACTGTATGTGTGCGTACGT........................................................................................................................ | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACTGTATGTGCGCATACGTGTGCTCA................................................................................................................. | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...ACCTGTATGTGTGCATACGT........................................................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............TGTGCATACGTGTGCTCATGC.............................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACG......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACGTTATGTGTGCATACGTGTGCTCA................................................................................................................. | 26 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCATACGCGTGCTCA................................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGCATACGTGTGCTTA................................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATGTGTGAATACGTGTGCTC.................................................................................................................. | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GTATGTGTGCATACGT........................................................................................................................ | 16 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GTATGTGTGCATACGTGTGCC................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACTGTATGTGTGCAT............................................................................................................................ | 16 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CGACTGTATGTGTGCATACGT........................................................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TGTGTGTGTGCATACGTGTG..................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................CACCTTAGAAATTCGTTGACTTTAAATC....................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...AACTTTATGTGTGCATACGT........................................................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGTGT...................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......TGTATATGTGCATACGTGTGCTCATGC.............................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACTGTATGTGTGCATACGTGTGA.................................................................................................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CCACTGTATGTGTGCATACGTGTGCT................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACTGTATGTGTGCATACGTGTGCTCAT................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chrU_M_105:846-1032 - | dsi_188 | AGGCAAGACAAACCAAGAATTTTTCGCATGGGGCCAATTAATCAAAAATATTATGGATTTAAAATCAACGAATTTCTAAGGTGAAGAGCATATTTTGTCAGATTTACAAGGCATGAGCACACGTATGCACACATACAGTTG-----------TA----------------------------------------------------------------------------------------------------- |
| droSec2 | scaffold_3:282005-282147 - | AATT----------------------------------------------------------------------------------------------------TACAATGCATGAGCACACGTATGCACACATACAGTTG-----------TATGCTATCACTGTATGCGTAGAAAAGAGCTGTTTGCTGTAGCGCTCTCCG-CTCTTTCTCTCTCGAACAAAAACTCAAGAGAGCCTGGAGCCACCTCTAGAG | |
| dm3 | chr3LHet:288350-288492 + | AATT----------------------------------------------------------------------------------------------------TACAATGCATGAGCATACGTGTGCACACATACAGTTG-----------TCTGCTATCACTGTATGCGTAGAAAAGAGCTGTTCGCTGTAGCGCTCTCCG-CTCTCTCGCTCTCTAACAAAAATTCGAGAGAGCCTGGAGCCACCTCTAGAG | |
| droEre2 | scaffold_4929:25135230-25135373 - | AATT----------------------------------------------------------------------------------------------------TACAATATATGAGCACACGTGTGCACACATACAGTGC-----------TCGGCTGTCGCTGTATGCGTAAAAGAGAGCTGCTGGCTCTAGAGCGCTCTCTCTCTCTGGCTCTTAAACAAAAATTCGAGAGAGCCTGGAGCCAGCTCTAGAG | |
| droYak3 | v2_chr2L_random_010:624168-624310 + | AATA----------------------------------------------------------------------------------------------------TACAAGGCATGAGCATACCCAGGCACACATACAGTGG-----------TCGGCTGTCACTGTATGCGTACAAGAAAGCTGCTGGCTCTAGAGCTTTCTG-CTCTCTGTGTTTTGAACAAAAATGCGAGAGAGCCTTGAGCCAGCTTTAGAG | |
| droBia1 | scf7180000301480:11888-12016 - | A---------------------------------------------------------------------------------------------------------CAATGCGCGAGAATACATGTGCACACATCCAATCG-----------TGTTCTGTCGCTGTATGCGTGCATGAGAGGCGCTAGCTCTAGAGCGCTCTC--TCTCTGGCTTTTGAGC--------AAGAGAGCCTGGAGCCAGCTCTAGAG | |
| droTak1 | scf7180000411080:3441-3574 + | CAAT----------------------------------------------------------------------------------------------------AACAATGCGCGAGAATACATGTGCACACATACAATCG-----------TCTTCTGTCTCTGTATGCGTGAATGAGAGCGGCTGGCTCTAGAGCGCTCTC--TCTCTGGCTTTTGAGC--------AAGAGAGCCTGGAGCCAGCTCTAGGG | |
| droRho1 | scf7180000776994:14800-14943 - | GTGCATTGCAGCGA------------------------------------------------------------------------------------------TGAAATGCTTGTGAATACATGTACACACATACAGTCG-----------TCGGCTGTAGCAGTATGCGTACAAGAGAGCGGCTGGCTCTAGAGCGCTTTC--TCGCTGGCTTTTGGAC--------AAGAGAGCGAAAAGCCACTTGAAAAG | |
| droKik1 | scf7180000302203:3047-3186 - | AAAT----------------------------------------------------------------------------------------------------TGCCCTTCTTGAGG-TCCGTTTGCACACAAACACCCG-----------GCTGC-GTCGCTGAATGCGTACAAGAGAACGGCTGGCTCTAGAGCGCTCTC-TTCGCTGGCTTTTGAACAA-AATTTAAGAGAGCCTGGAGCCAGTTCAAAAG | |
| dp5 | 4_group3:9794423-9794559 - | GATC----------------------------------------------------------------------------------------------------TCCAATGCATGAGCATACGTGTGCACACACACAGTCATATCTCAGTCCTCGGCTGCCTCATGACGAACACGAAAGAGCGGCTAGCTTTTGAGTGCTCTC--GCTCCTTCTCTCA-----GAAGGCAGAAGAGCAAGAAGC----------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
Generated: 12/06/2013 at 02:33 PM