

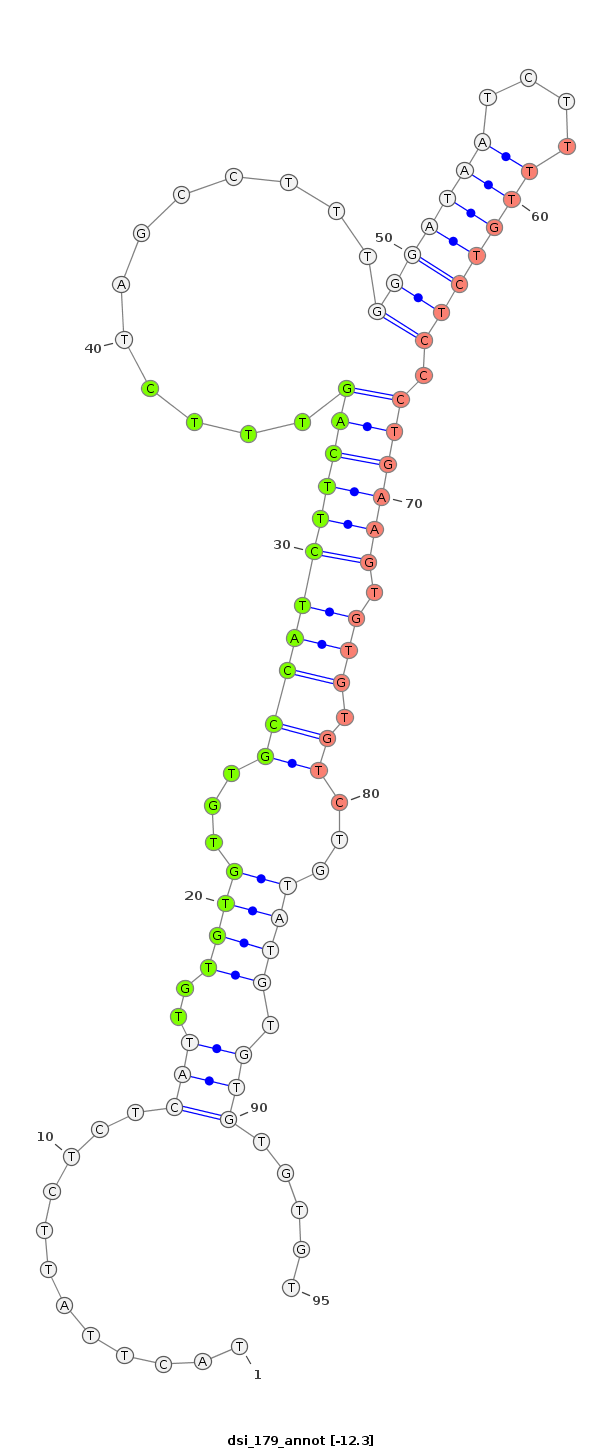
ID:dsi_179 |
Coordinate:chrU_M_165:3515-3586 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
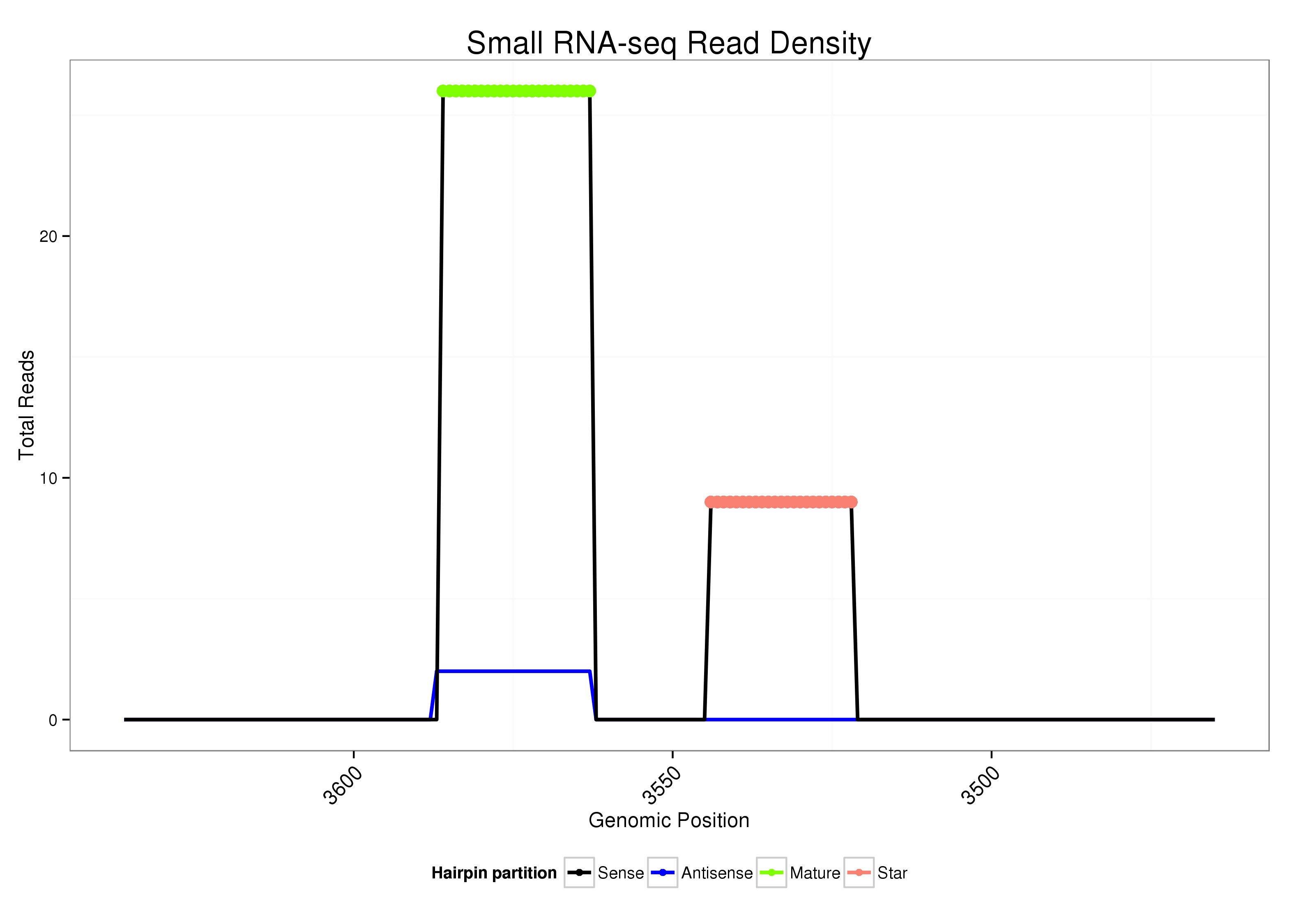
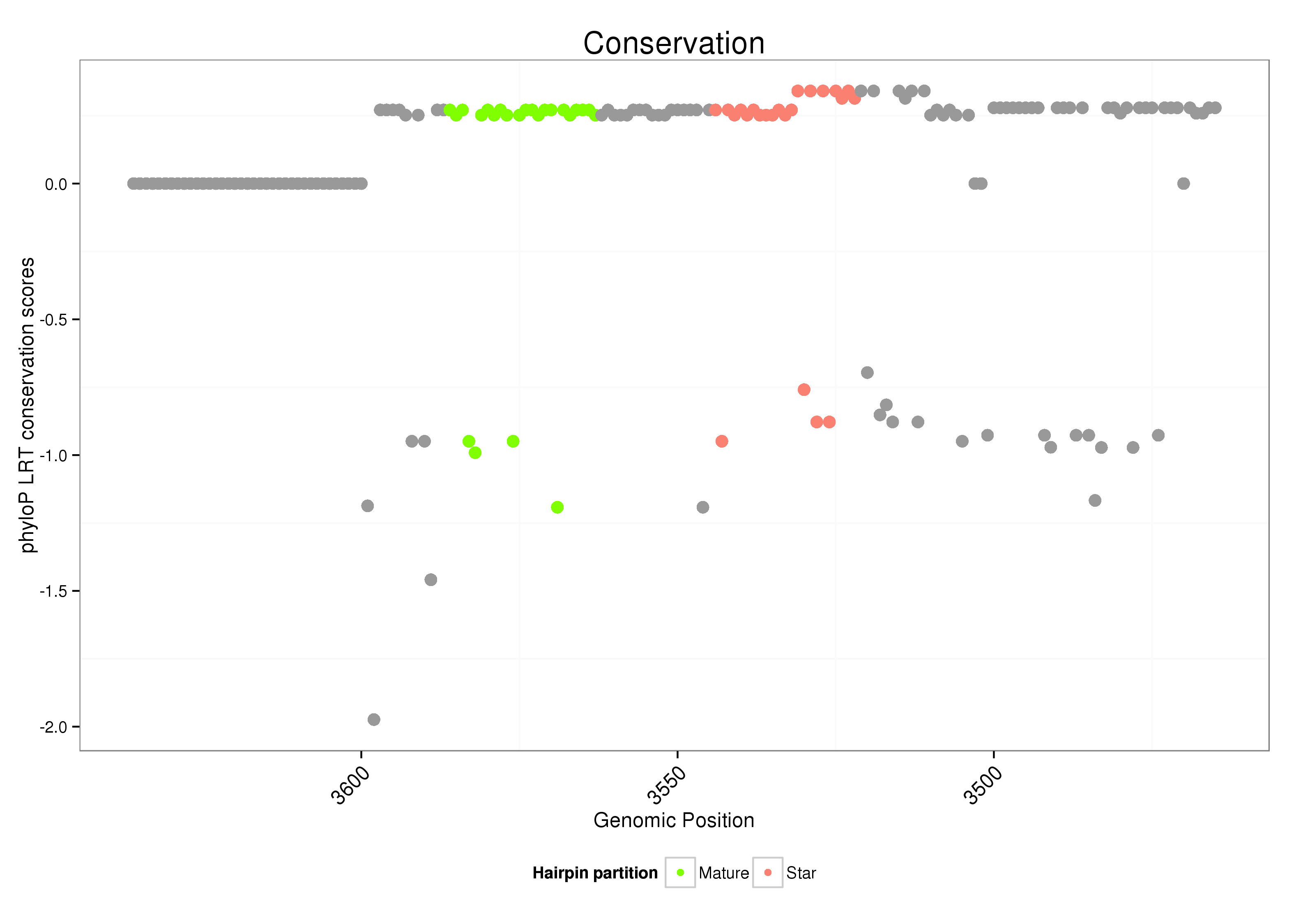
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -12.3 | -12.3 | -12.3 |
 |
 |
 |
intergenic
No Repeatable elements found
|
NNNNNNNNNNNNNNNNNCGCCACTGTGGTGGATTCTACTTATTCTCTCATTGTGTGTGTGCCATCTTCAGTTTCTAGCCTTTGGGATAATCTTTTGTCTCCCTGAAGTGTGTGTCTGTATGTGTGTGTGTGGGCTAATTAATATTAAATATTTTTTGATTTTGATTGTCGAT
***********************************........................(((((...(((......(((((((((((((....)))))))...)))).))....)))...))).))....****************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR618934 Ovary |
SRR553487 Ovary |
O002 Head |
SRR553485 Ovary |
SRR553488 Ovary |
SRR553486 Ovary |
M025 embryo |
M053 Female-body |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TGTGTGTGTGCCATCTTCAGTTTC.................................................................................................. | 24 | 0 | 5 | 26.00 | 130 | 125 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TTTGTCTCCCTGAAGTGTGTGTC......................................................... | 23 | 0 | 20 | 9.35 | 187 | 187 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TGTGTCTGTATGTGTGTGTGT.......................................... | 21 | 0 | 20 | 0.80 | 16 | 0 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TGTGTGTCTGTATGTGTGT.............................................. | 19 | 0 | 20 | 0.75 | 15 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGTGTGTGTGCCATCTTCAGTGCCTGGCC............................................................................................. | 29 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................TCTCCCTGAAGTGTGTGTCTGTATG................................................... | 25 | 0 | 20 | 0.45 | 9 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTTTGTCTCCCTGAAGTGTGTGTC......................................................... | 24 | 0 | 20 | 0.30 | 6 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................CCTGAAGTGTGTGTCTGTATGTGTG............................................... | 25 | 0 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TTAAATATTTTTTGATTTTGATTGTCGAT | 29 | 0 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCCCTGAAGTGTGTGTC......................................................... | 17 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TACTTATTCTCTCATTGTGTGTG.................................................................................................................. | 23 | 0 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| ..................................................................................................TCCCTGAAGTGTGTGTCTGTATGTGT................................................ | 26 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ...............................................................................TTTGGGATAATCTTTTGTCT......................................................................... | 20 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TGTGTGTCTGTATGTGTGTGTG........................................... | 22 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................TGTGTGGGCTAATTAATATTAAATA...................... | 25 | 0 | 20 | 0.10 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TTGTCTCCCTGAAGTGTGTGTCTGTATG................................................... | 28 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TTTGTCTCCCTGAAGTGTG............................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TGAGTGTCTGTATGTGTGT.............................................. | 19 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TGTCTGTATGTGTATGTGTGGGCTA.................................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTTTGTCTCCCTGAAGTGTGT............................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................GTCTGTATGTGTGTGTGTTGG....................................... | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................GTGGGCTAATTAATATTAAATATTTTT................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................AGTGTGTGTCTGTATGTGTG............................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................TGTGTCTGTATGTGTGTGTGTG......................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................TGGGCTAATTAATATTAAATATTTTT................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................CTCTCATTGTGTGTGTGCCA............................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................TCTTTTGTCTCCCTGAAGTGTGTGTCT........................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TAATCTTTTGCCTCCCTGAAGTGTG............................................................. | 25 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTTTGTCTCCCTGAAGTGTGTG........................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................TGTGTGGGTTAATTAATATTAAATA...................... | 25 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TTTGTCTCCCTGAAGTGTGTGTCTGTAA.................................................... | 28 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TTTGTCTCCCTGAAGTGTGTCGC......................................................... | 23 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTTTGTCTCCCTGAAGTGTGTGTCTG....................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
ATCGACAATCAAAATCAAAAAATATTTAATATTAATTAGCCCACACACACACATACAGACACACACTTCAGGGAGACAAAAGATTATCCCAAAGGCTAGAAACTGAAGATGGCACACACACAATGAGAGAATAAGTAGAATCCACCACAGTGGCGNNNNNNNNNNNNNNNNN
******************************************....((.(((...(((....((.((((...(((((((....)))))))))))))......)))...)))))........................*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Ovary |
SRR553486 Ovary |
SRR618934 Ovary |
SRR553488 Ovary |
SRR553487 Ovary |
M053 Female-body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................CACATACAGACACACACTTCAGGGA.................................................................................................. | 25 | 0 | 20 | 2.65 | 53 | 53 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TAAGTAGTATCCATTACAGTGGCG................. | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................AATGATAGAAGATGTAGAAT............................... | 20 | 3 | 12 | 0.75 | 9 | 8 | 0 | 0 | 0 | 1 | 0 |
| ................................................ACACATACAGACACACACTTCAGGGA.................................................................................................. | 26 | 0 | 20 | 0.55 | 11 | 11 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CACACACACAATGAGAGAATAAGT.................................... | 24 | 0 | 20 | 0.45 | 9 | 8 | 0 | 1 | 0 | 0 | 0 |
| .....................................................TACAGACACACACTTCAGGGAGACAAA............................................................................................ | 27 | 0 | 20 | 0.30 | 6 | 0 | 0 | 0 | 5 | 1 | 0 |
| .................................................................................GATTATCCCAAAGGCT........................................................................... | 16 | 0 | 20 | 0.30 | 6 | 0 | 0 | 6 | 0 | 0 | 0 |
| .........................................................GACACACACTTCAGGGA.................................................................................................. | 17 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCATACAGACACACACTTCAGGGA.................................................................................................. | 24 | 1 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TACAGACACACACTTCAGGGAGACAAAA........................................................................................... | 28 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .............................TATTAATTAGCCCACACACACACATA..................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................CACACATACAGACACACACTTCAGGG................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TACAGACACACACTTCAGGG................................................................................................... | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..................................................ACATACAGACACACACTTCAGGGAGACAAAA........................................................................................... | 31 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACATACAGACACACACTTCAGTGA.................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CACACACACAATGAGAGA.......................................... | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................TGGCACACACACAATGAGAGAATA....................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GACACACACTTCAGGGAGACAAAAGA......................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................CACACACTTCAGGGAGACAAAAGATT....................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................TTAGTATTAATTAGACCA................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................CACACACATACAGACACACACTTCAGGGA.................................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................ACACACATACAGACACACACTTCAGGGA.................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................TACAGACGCACACTTCAGGGAAACAAAA........................................................................................... | 28 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................CACATACAGACACACACTTCAGGGAGAC............................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................CACATACAGACACACACTTCAGGGAG................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............TCAAAAAATATTTAATATTAATTAGCCC.................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACGTACAGACACACACTTCAGGGAGA................................................................................................ | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACGTACAGACACACACTTCAGGGA.................................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................CACATACAGACACACACTTCAGGGAGA................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................ACACATACAGACACACACTTCAGGGAG................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................TTAATTAGCCCACACACACACATACAGA................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................CACACATACAGACACACACTTCAGGGAG................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................TAAAGACACACACTTCAGGGAGACAAAAGA......................................................................................... | 30 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................CACACACATACAGACACACACTTCAGGGAA................................................................................................. | 30 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................ACACACTTCAGGGAGAC............................................................................................... | 17 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...GACAATCAAAATCAAAAAATATTTAATA............................................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................TAATATTAATTAGCCCACACACACACA....................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chrU_M_165:3465-3636 - | dsi_179 | NNNNNNNNNNNNNNNNNCGCCACTGTGGTGGATTCTACTTATTCTCTCATTGTG------TGTGTGCCATCTTCAGTTTCTAG-CCTTTGGGATAATCTTTTGTCTCCCTGAAGTGTGTGTCTGT------------ATGTGTGTGTGTGGGCTAATTAATATTAAATATTTTTTGATTTTGATTGTCGAT |
| droSec2 | scaffold_4173:2119-2257 + | -----------------------------------TACTTATTCTCTCATTGTG----TGTGTGTGCCATCTTCAGTTTGCAG-CCTTTGGGATAATCTTTTGTCTCCCTGAAGTGTGTGTCTGT------------ATGTGTGTGTGTGGGCTAATTAATATTAAATATTTTTTGATTTTGATTGTCGAT | |
| dm3 | chrX:6059944-6060043 + | -------------------------------------CTTATTCACCCATTGTGTGTGTGTGTGTGTCATCTTCAGCTTGCAG-CCTTTGGCATAATCTTTTGTCTCCCTGATGTGTGTGTCTGT------------ATGTGTGTGTGTG----------------------------------------- | |
| droEre2 | scaffold_4690:15038761-15038871 + | -------------------------------------CCTATTCCCCGATTGTA----TGAGTGTGTCATCTTAAGTTTGCAGGCCTTTGGGATAATATTCTGTCTCCCTGAAGTGTGTGTCTATCTTTCTATCTTTGTGTGTGTGTGTGAG--------------------------------------- | |
| droYak3 | v2_chrUn_3925:663-721 + | -------------------------------------GA-------------------------------------------------------------------------AATTTTTGTCTAT------------AATTGTTT---------GATTAATATCTAATGTCGATTGAATTTAATT-TCGAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 12/06/2013 at 02:33 PM