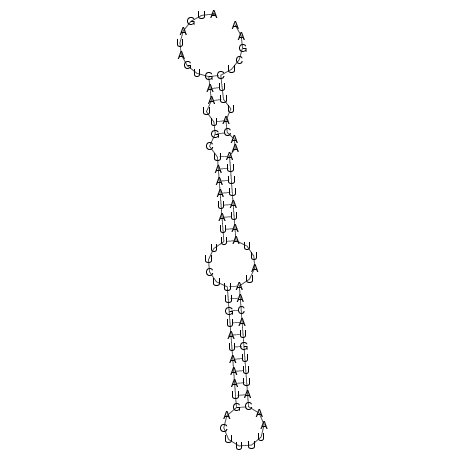
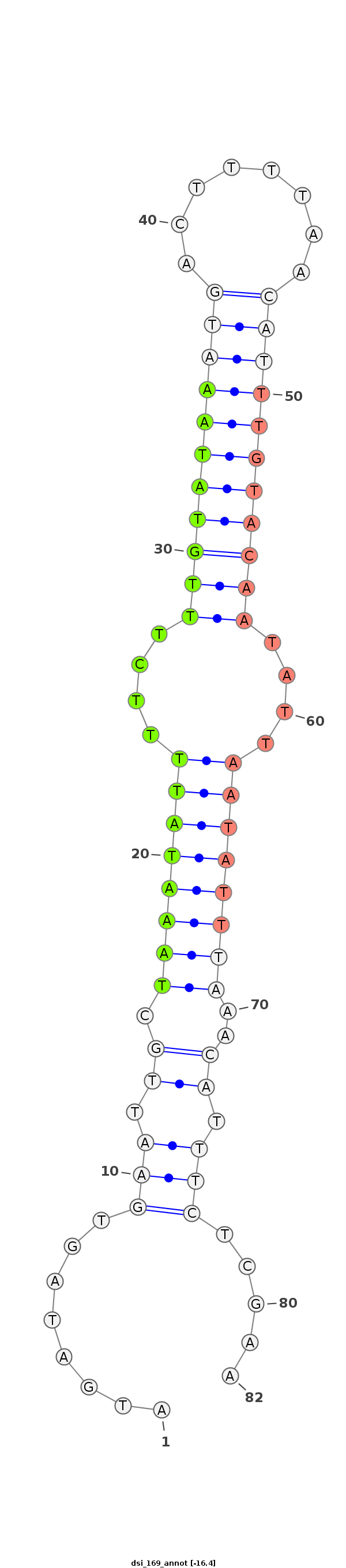
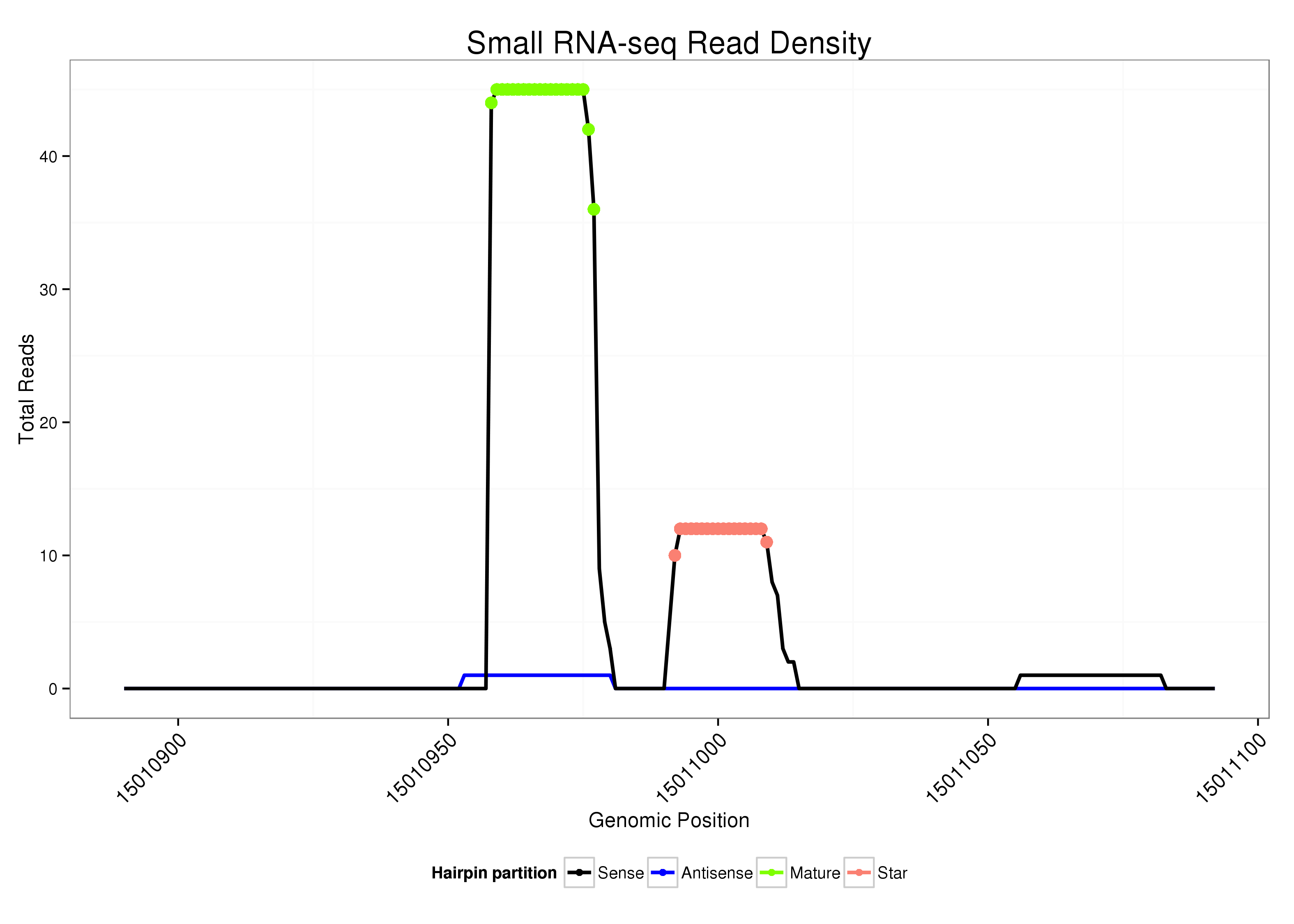
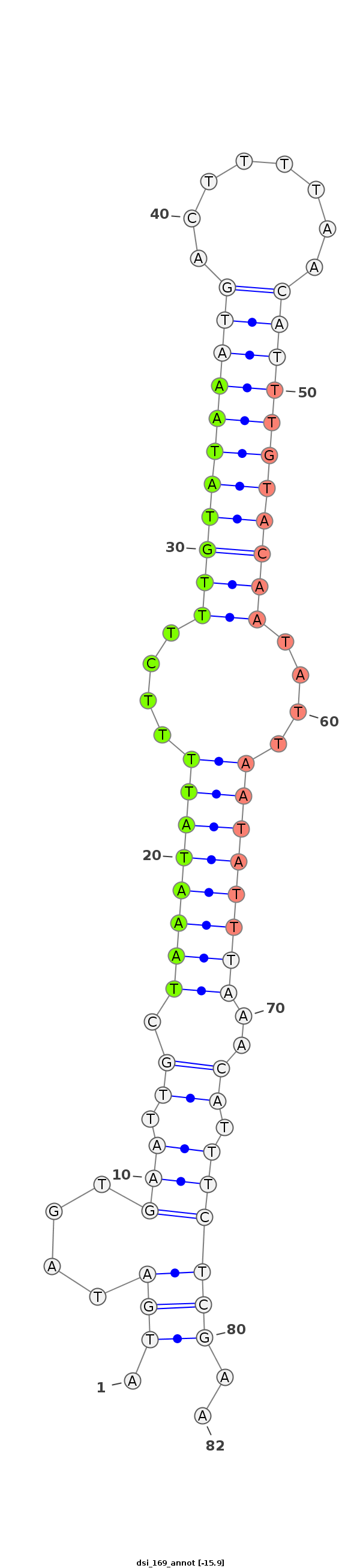
ID:dsi_169 |
Coordinate:X:15010940-15011042 + |
Confidence:Known Ortholog |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -15.9 |
 |
intergenic
No Repeatable elements found
|
AACAACAATTCAAATGCAAGCAAAAAAAATAAAAAATAAAAAAGCTGAAGAGAATGATAGTGAATTGCTAAATATTTTCTTTGTATAAATGACTTTTAACATTTGTACAATATTAATATTTAAACATTTCTCGAATCAAAGTATGCCACATCAGCTTTGCAAGCCAAACGTGCGATGGACTAGTGACGAAAGACACTAAAAGT
*****************************************************........(((.((.((((((((....(((((((((((........)))))))))))....))))))))..)).))).....******************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M024 male body |
M023 head |
O002 Head |
SRR553488 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................TAAATATTTTCTTTGTATAA................................................................................................................... | 20 | 0 | 1 | 26.00 | 26 | 25 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTATA.................................................................................................................... | 19 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTATAAA.................................................................................................................. | 21 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTATAAATG................................................................................................................ | 23 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................TTGTACAATATTAATATT................................................................................... | 18 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTAT..................................................................................................................... | 18 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TTTGTACAATATTAATATTTAAAC.............................................................................. | 24 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTATAAAT................................................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................TTGTACAATATTAATATTTA................................................................................. | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................AACGTGCGATGGACTAGTGACGAAAGA.......... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................TGTACAATATTAATATTTAA................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TTTGTACAATATTAATAT.................................................................................... | 18 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TGTACAATATTAATATTTA................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................TTTGTACAATATTAATATTT.................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................TTTGTACAATATTAATATTTA................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TAAATATTTTCTTTGTATAAAA................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................AAATATTTTCTTTGTATAA................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................ATTTGTATAAATGACTT............................................................................................................ | 17 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................ATTTTCTTTGTACGTATGACT............................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................ATAAAAAAGGTGAAGA........................................................................................................................................................ | 16 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................ATCAAAGTATGTCAGAT.................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................AGTGACGAATGACACCA..... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ACTTTTAGTGTCTTTCGTCACTAGTCCATCGCACGTTTGGCTTGCAAAGCTGATGTGGCATACTTTGATTCGAGAAATGTTTAAATATTAATATTGTACAAATGTTAAAAGTCATTTATACAAAGAAAATATTTAGCAATTCACTATCATTCTCTTCAGCTTTTTTATTTTTTATTTTTTTTGCTTGCATTTGAATTGTTGTT
********************************************************************.....(((.((..((((((((....(((((((((((........)))))))))))....)))))))).)).)))........***************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 Ovary |
|---|---|---|---|---|---|---|
| ...............................................................TTTGATTCGAGAAATGTTTAAATATTAA................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 |
| .............................................................................................TTGTACAAGTGTTAAAA............................................................................................. | 17 | 1 | 5 | 0.20 | 1 | 0 |
| ............................................................................................................................................................................TATTTTTTTTAATTGCAGTTGAA........ | 23 | 3 | 7 | 0.14 | 1 | 0 |
| .............................................................................................................................................................AGCTGTATTATTTTTTATTT.......................... | 20 | 2 | 16 | 0.06 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | X:15010890-15011092 + | dsi_169 | AACAACA--------------ATTCAAATGCAAGCAA----AAAAAATAAAAAATAAAAAAGCTGAAGAGAATGATAGTGA-ATTGCTAAATATTTTCTTTGTATAAATGACTTTTAACATTTGTACAATATTAATATTTAAACATTTCTCGAATCAAAGTATGCCACATCAGCTTTGCAAGCCAAACGTGCGATGGACT----AGTGACGAAAGACACTAAAAGT |
| droSec2 | scaffold_8:1727558-1727761 + | AACAACA--------------ATTCAAATGCAAGCAATCTTAAAAA---AAAAATAAAAAAGCTGAAAAGAATGATAGTGA-ATTGCTAAATATTTTCTTTGTATAAATAACTTTTAATATTTGTACAATATTAATATTTAAACATTTCTCGAATCAAAGTATGCGACATCAGCTTTGCAAGCCAAACGTGCGATGGACT----AGTGACGAAAGACACTAAAAGT | |
| dm3 | chrX:19445123-19445329 + | dme-mir-972 | AAAAATACAGCAAATAGGGAGATTCAAACCCAT---ATTTGAAAAA---AAAATGCAAATAGCTAAAGAGAATGATAGGGAAATTGCTAAATATTTTTTTTGTATAAATAACTTTTAACTTTTGTACAATACGAATATTTAGGCATTTCTCAAATCAAA-------------GATTTGCAAGCCAAACGTGCAATGAACTACATAGTTACGAAAGACACTAAAAGT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 12/06/2013 at 02:33 PM