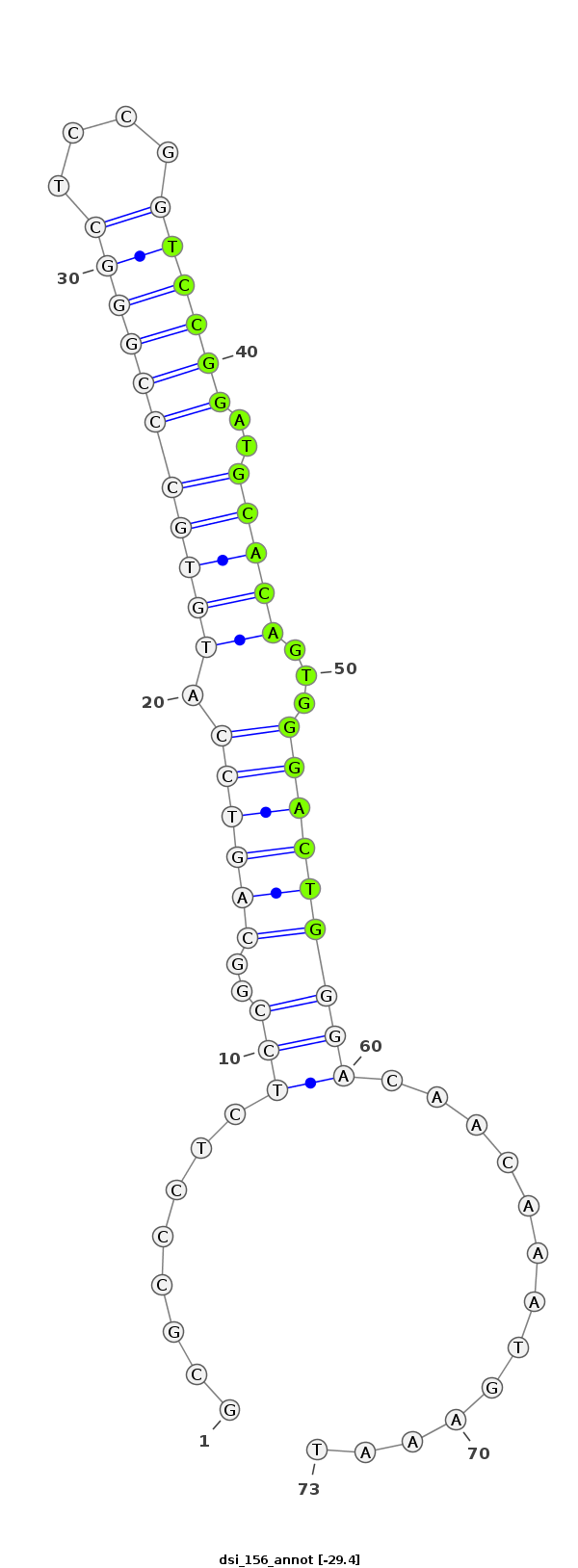
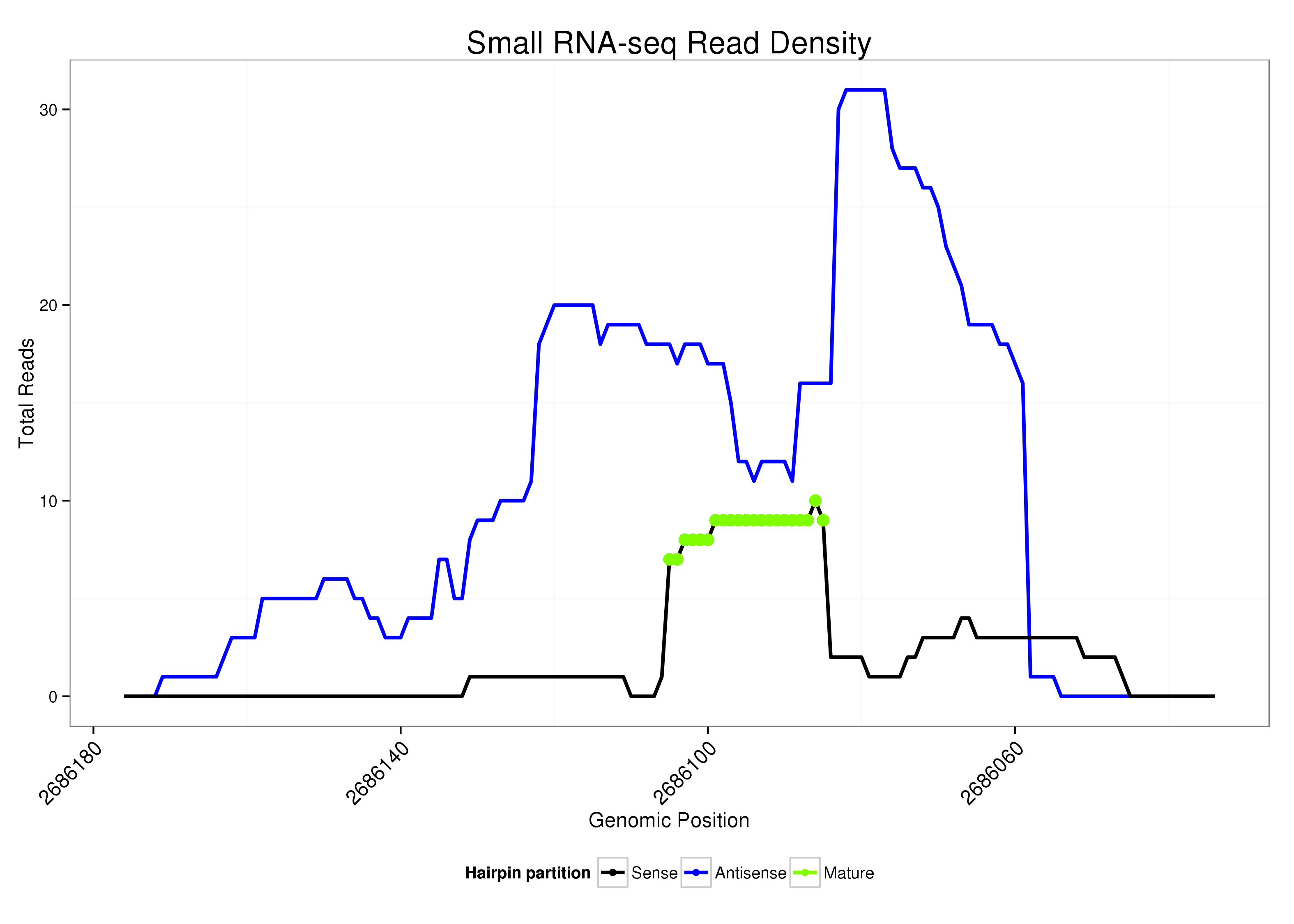
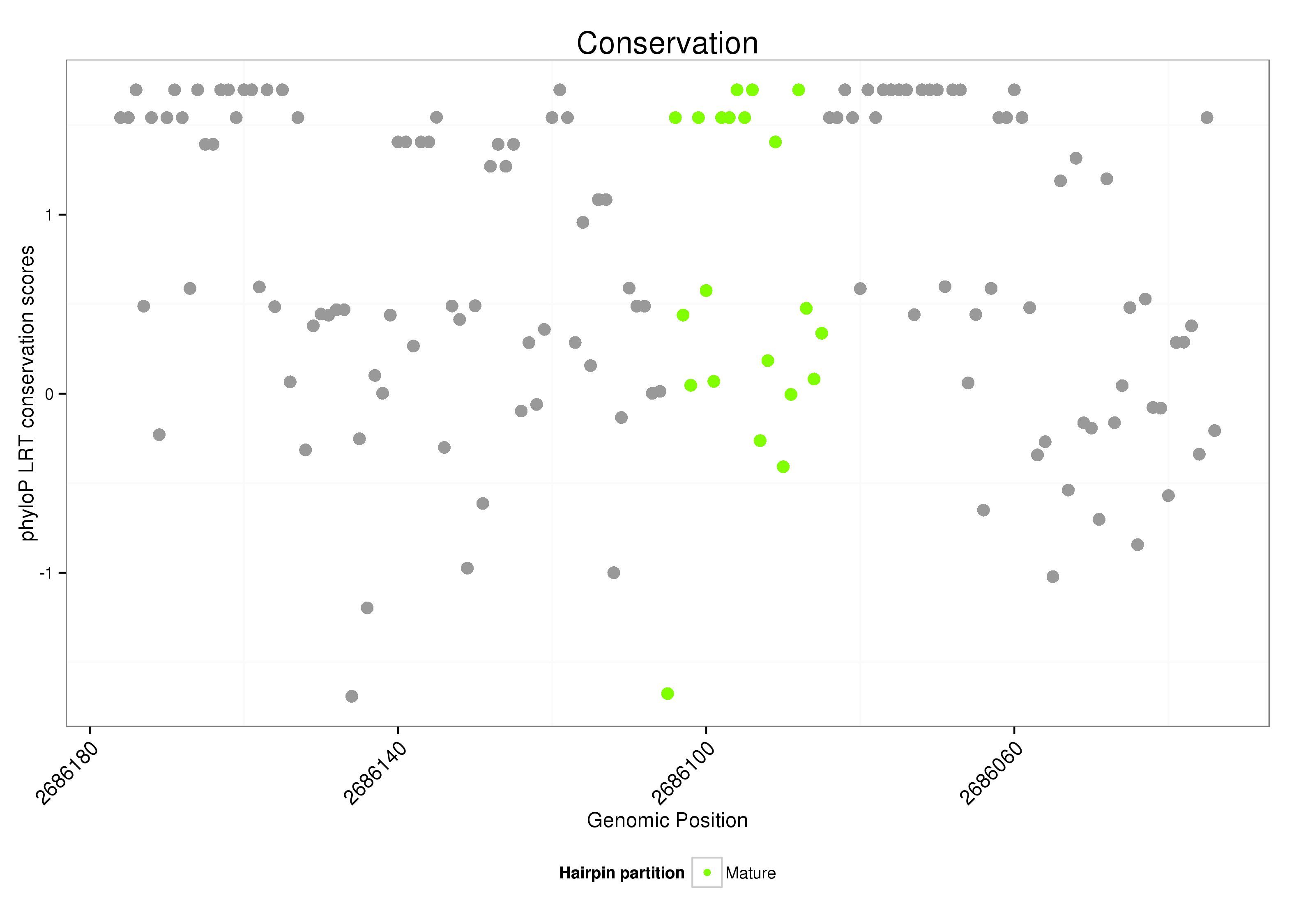
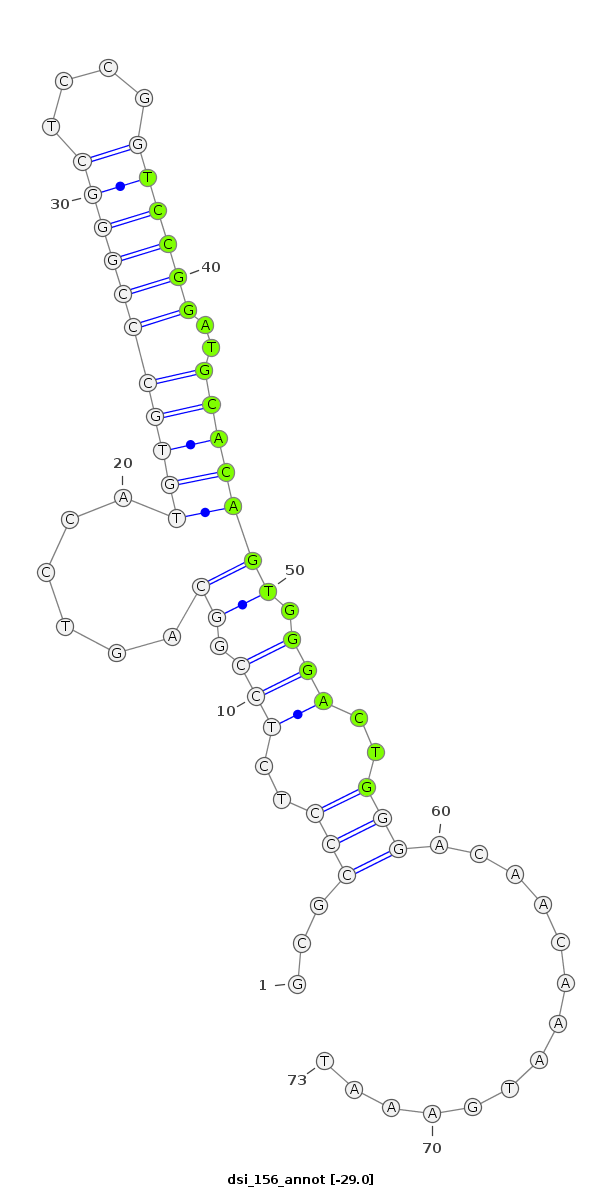
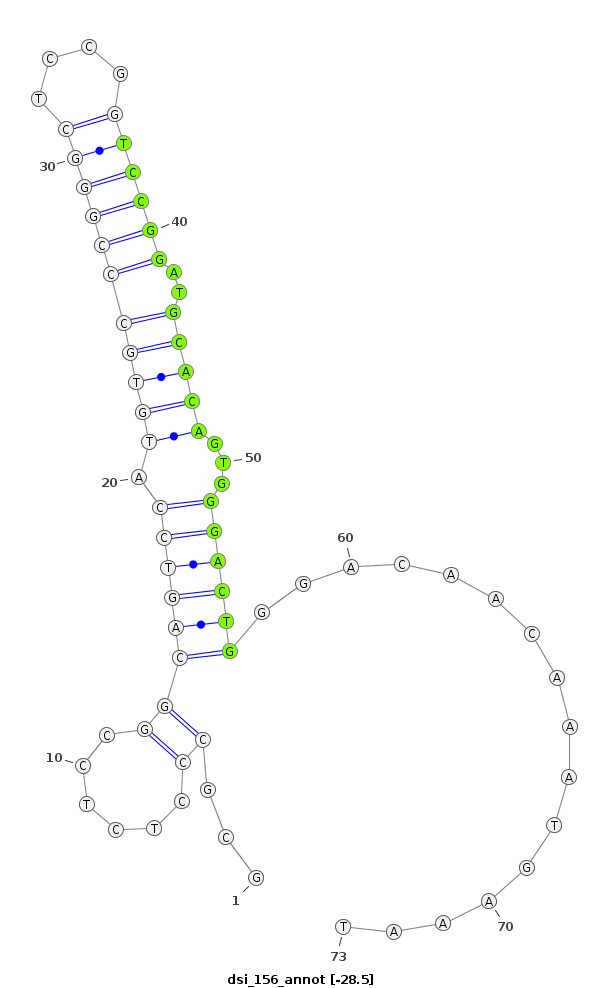
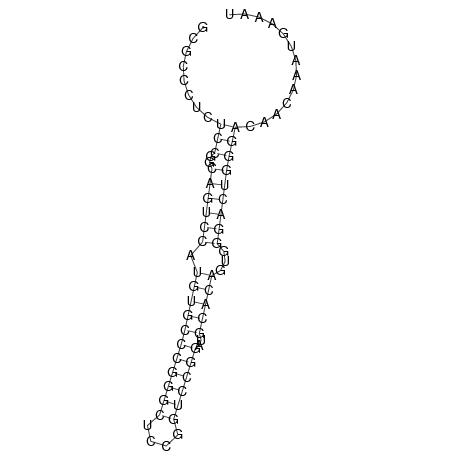| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCGC......................... |
25 |
0 |
1 |
14.00 |
14 |
14 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................TCCCACTGTGCATCCGGACCGGAGCC............................................................... |
26 |
0 |
1 |
5.00 |
5 |
1 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................GAGGGCGCCATGTGGCGCAA....................... |
20 |
2 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................CGGGCACATGGACTGCCGGAGAGGGCGCCA................................. |
30 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................CGCGGGCACATGGACGGACGGA........................................... |
22 |
3 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| .............................................TTGTCCCAGTCCCACTGTGCA............................................................................. |
21 |
0 |
1 |
2.00 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ............................................................................................CTGCCGGAGAGGGCGCCATGTGGCGCA........................ |
27 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................TCCGGACCGGAGCCCGGGCACATGGAC.................................................. |
27 |
0 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................GGAGCCCGGGCACATGGACTGCCGGA........................................... |
26 |
0 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| ..................TCACGAACTGCACCCAAATTTCATT.................................................................................................... |
25 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................TTTGTTGTCCCAGTCCCACTG................................................................................. |
21 |
0 |
1 |
2.00 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCGCA........................ |
26 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................CGCGGGCACATGGACGGACG............................................. |
20 |
3 |
6 |
1.50 |
9 |
0 |
0 |
9 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................CGCGGGCACATGGACGGACGG............................................ |
21 |
3 |
3 |
1.33 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
| .....................................TTCATTTGTTGTCCCAGTCCCACTG................................................................................. |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..........................TGCACCCAAATTTCATTTGTTGTC............................................................................................. |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ......................................................TCCCACTGTGCATCCGGACCGGAGCCCG............................................................. |
28 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................CGGAGCCCGGGCACATGGACTGCCGGA........................................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGCGCCATGTGGC........................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................GCATCCGGACCGGAGCC............................................................... |
17 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................TTGTCCCAGTCCCACTGTGCATCCGGACC..................................................................... |
29 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................GCACATGGACTGCCGGAGAGG....................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGCG.................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................CGGGCACATGGACTGCCGGAGAGGGCGC................................... |
28 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................TGTCCCAGTCCCACTGTGCATCCGGA....................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGCGCCATGTG............................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............GCGCTCACGAACTGCACC............................................................................................................... |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................CGGGCACATGGACTGCCGGAGAGGGCG.................................... |
27 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................TCCCACTGTGCATCCGGACCGGAGC................................................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................CCCAGTCCCACTGTGCATCCGGA....................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCGCAA....................... |
27 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCGA......................... |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............CGCGCTCACGAACTGCACCCA............................................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................GTCCCACTGTGCATCCGGACCGGAGC................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGCGCC.................................. |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................CCACTGTGCATCCGGACCGG................................................................... |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................CCGGAGCCCGGGCACATGGACTGCCGGAG.......................................... |
29 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....GAAGAAAGCGCGCTCACGAACTGCA................................................................................................................. |
25 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................TTTGTTGTCCCAGTCCCACTGTGCATC........................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................TGCATCCGGACCGGAGCCCGGGCAC........................................................ |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .......................................................CCCACTGTGCATCCGGACC..................................................................... |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGC..................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGGACTGCCGGAGAGGGCGCCATGGG............................. |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCG.......................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................CCAGTCCCACTGTGCATCCGGACCGGAGCC............................................................... |
30 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................GCCGGAGAGGGCGCCATGTGGCGC......................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................TGCCGGAGAGGGCGCCATGTGGCGCCGGA..................... |
29 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................CACATGGACCGTCGGCGAGG....................................... |
20 |
3 |
5 |
0.80 |
4 |
0 |
0 |
0 |
2 |
0 |
1 |
1 |
0 |
| ....................................................................................CACATGGACCGTCGGCGAG........................................ |
19 |
3 |
15 |
0.53 |
8 |
0 |
0 |
0 |
2 |
0 |
5 |
1 |
0 |
| ................................................................................CGGGCACATGGACGGACGGA........................................... |
20 |
2 |
6 |
0.50 |
3 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................CACATGGACCGTCGGCGAGGG...................................... |
21 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................ACATGGACCGTCGGCGAGG....................................... |
19 |
3 |
17 |
0.18 |
3 |
0 |
0 |
0 |
0 |
0 |
3 |
0 |
0 |
| .............................................................................................................ATGGGGCGCCGGAGA................... |
15 |
1 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........AAGGCGCCCTGACGAACTGC.................................................................................................................. |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GCGGGCACATGGACGGACGGA........................................... |
21 |
3 |
10 |
0.10 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................TGAGAGCGCGCCATGTG............................. |
17 |
2 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................CCGGAGCCGGGGCAC........................................................ |
15 |
1 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TATCGGAACCGGAGCCCGG............................................................ |
19 |
3 |
16 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................CGATGTGGGGACGGAGAAT................. |
19 |
3 |
17 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................CTGGCACATGGACGGACGGA........................................... |
20 |
3 |
18 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |