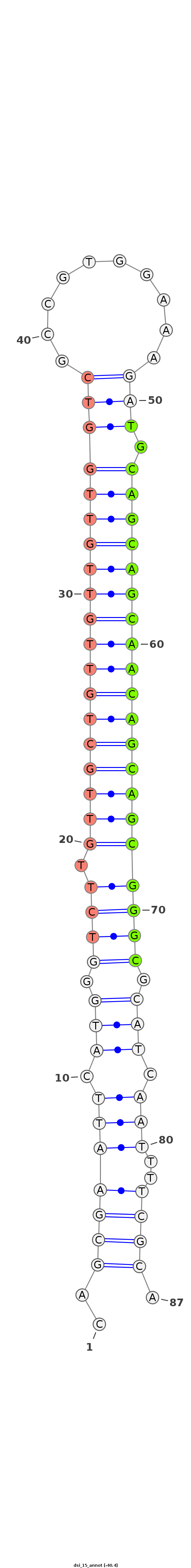
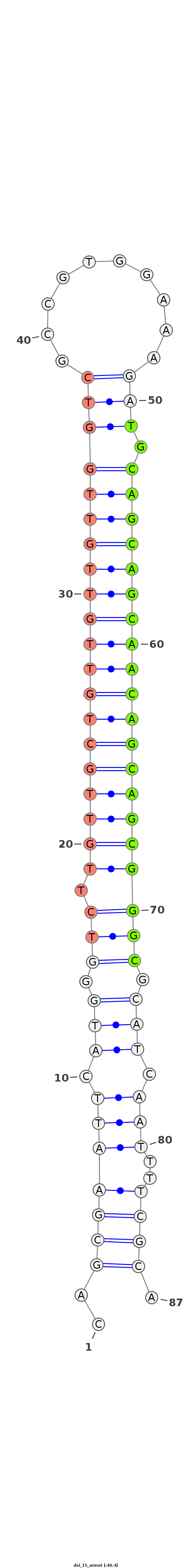
| .....................................................................................TGCAGCAGCAACAGCAGCGGGC................................................... |
22 |
0 |
2 |
359.00 |
718 |
548 |
108 |
6 |
52 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCG.................................................. |
23 |
0 |
2 |
54.50 |
109 |
43 |
31 |
0 |
7 |
17 |
7 |
0 |
2 |
2 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGTC..................................................................................... |
23 |
0 |
2 |
52.00 |
104 |
37 |
54 |
2 |
5 |
4 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGG.................................................... |
21 |
0 |
2 |
19.50 |
39 |
0 |
0 |
8 |
0 |
6 |
3 |
8 |
7 |
1 |
3 |
3 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCT.................................................. |
23 |
1 |
2 |
18.50 |
37 |
33 |
3 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGG..................................................... |
20 |
0 |
3 |
14.00 |
42 |
0 |
0 |
33 |
0 |
0 |
1 |
6 |
1 |
1 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGAC................................................... |
22 |
1 |
2 |
13.50 |
27 |
23 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCG...................................................... |
19 |
0 |
3 |
13.00 |
39 |
0 |
0 |
39 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGG....................................................................................... |
21 |
0 |
2 |
5.00 |
10 |
6 |
0 |
2 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGT...................................................................................... |
22 |
0 |
2 |
4.00 |
8 |
3 |
3 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGTCT.................................................................................... |
24 |
1 |
2 |
3.00 |
6 |
4 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................CAGCAGCAACAGCAGCGGGC................................................... |
20 |
0 |
2 |
3.00 |
6 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................GGTCTTGTTGCTGTTGTTGTTGG....................................................................................... |
23 |
0 |
2 |
2.50 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGT................................................... |
22 |
1 |
2 |
2.50 |
5 |
1 |
1 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGA................................................... |
22 |
1 |
3 |
2.33 |
7 |
2 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
1 |
2 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCGC................................................. |
24 |
0 |
2 |
2.00 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCTT................................................. |
24 |
2 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTTGTC..................................................................................... |
23 |
1 |
2 |
1.50 |
3 |
0 |
0 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................CTTGTTGCTGTTGTTGTTGGTC..................................................................................... |
22 |
0 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGG................................................... |
22 |
1 |
3 |
1.33 |
4 |
3 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTTTTGGTC..................................................................................... |
23 |
1 |
2 |
1.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTT......................................................................................... |
19 |
0 |
4 |
1.00 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTG........................................................................................ |
20 |
0 |
3 |
1.00 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCATCGGGC................................................... |
22 |
1 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................GCAGCAGCAACAGCAGCGGGCG.................................................. |
22 |
0 |
2 |
1.00 |
2 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................ATAATGCCGGCGACAGCGAA.................................................................................................................... |
20 |
0 |
2 |
1.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCATCAGCAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
1.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGAC..................................................................................... |
23 |
1 |
2 |
1.00 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................GCAGCAACAGCAGCGGGC................................................... |
18 |
0 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGAAGCGGGC................................................... |
22 |
1 |
2 |
1.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCC.................................................. |
23 |
1 |
3 |
1.00 |
3 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCTA................................................. |
24 |
2 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................TCAGCAGCAACAGCAGCGGGC................................................... |
21 |
1 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................TGTTGCTGTTGTTGTTGGTC..................................................................................... |
20 |
0 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGTCC.................................................................................... |
24 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCCGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTATTGGTC..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TCCAGCAGCAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTTTTGCTGTTGTTGTTGGTC..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................AGCAGCAACAGCAGCGGGCGCA................................................ |
22 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCATCGGGCG.................................................. |
23 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................GAATTCATGGGTCTTGTTGCTG................................................................................................. |
22 |
0 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................AGCAGCAACAGCAGCGGGC................................................... |
19 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGGTGTTGTTGGTC..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................GCAACAGCAGCGGGCG.................................................. |
16 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTTTTGTTGGTC..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGAAGCAGCAACAGCACCGGGC................................................... |
22 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTTTTTGTC..................................................................................... |
23 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCGAT................................................ |
25 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGGC..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGTCGC................................................................................... |
25 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCATCAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................AGCAACAGCAGCGGGCG.................................................. |
17 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGCAGCAACAGCAGCGGGC................................................... |
19 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCACCGGGCG.................................................. |
23 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGTAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGTT..................................................................................... |
23 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TGCAGCAACAGCAGCGGGCG.................................................. |
20 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................GGTCTTGTTGCTGTTGTTGTTG........................................................................................ |
22 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCGT................................................. |
24 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCTGCAGCAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCGACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGG...................................................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCACCAGCAACAGCAGCGGGC................................................... |
22 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................TTGTTGCTCTTGTTGTTGGTC..................................................................................... |
21 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTTTTGGT...................................................................................... |
22 |
1 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGTTGTTGTTGTTGGTC..................................................................................... |
23 |
1 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTTTTGG....................................................................................... |
21 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGGG.................................................. |
23 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCGGCGGGCT.................................................. |
23 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTTG....................................................................................... |
21 |
1 |
7 |
0.29 |
2 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCACCCGCCGCGGGCG.................................................. |
23 |
3 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCTGCAAGGGCAGCGGGCG.................................................. |
23 |
3 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGA...................................................................................... |
22 |
1 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTCGTTGTTTGTC..................................................................................... |
23 |
2 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAACGGGC................................................... |
22 |
1 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GGCAGCAGCAACAGCAGCGGGC................................................... |
22 |
1 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTGTTGTTGGC...................................................................................... |
22 |
1 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................CCTTGTTGCTGTTGTTGTTGGT...................................................................................... |
22 |
1 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACATCAGCGGAC................................................... |
22 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGGCTAT................................................ |
25 |
3 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGT.................................................... |
21 |
1 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................................ACAGCAGCGGCCATATCAATT.......................................... |
21 |
3 |
10 |
0.10 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TCTTGTTGCTGTTTTTTTTTGTC..................................................................................... |
23 |
3 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................AGCAGCAACAGCAGC....................................................... |
15 |
0 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................TGCAGCAGCAACAGCAGCGGAT................................................... |
22 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................GCTACAGGAACGACCAG....... |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |