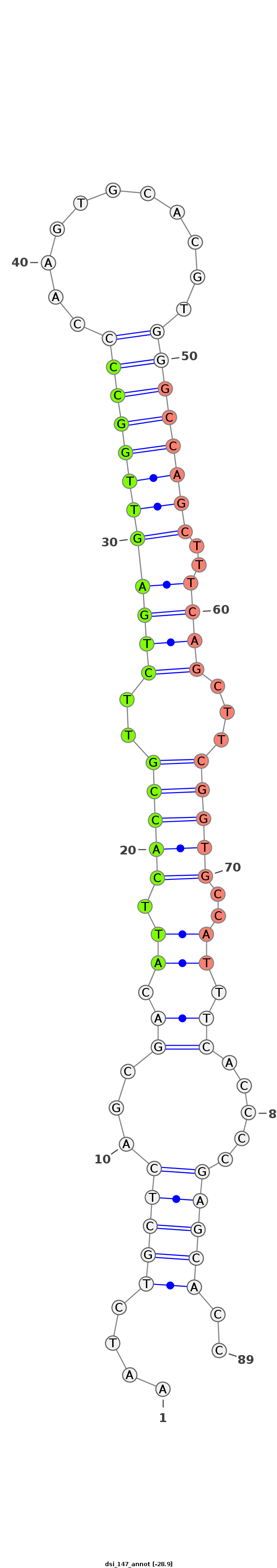
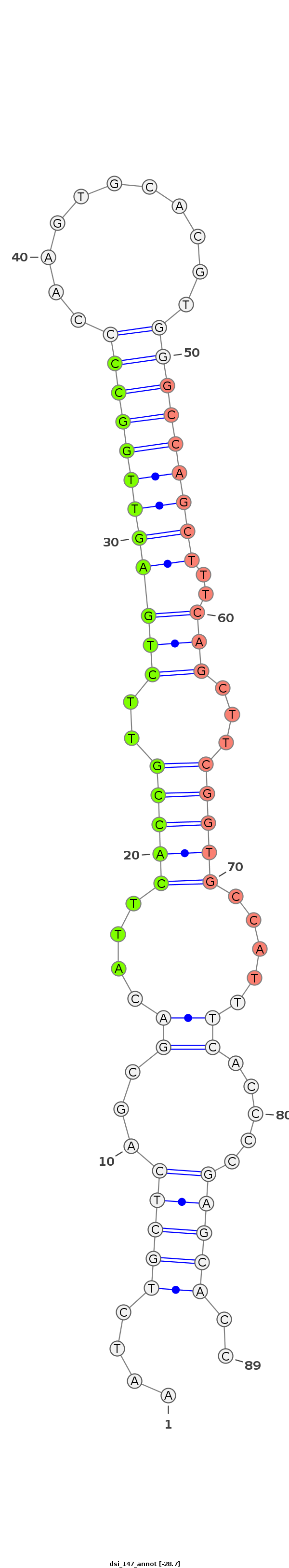
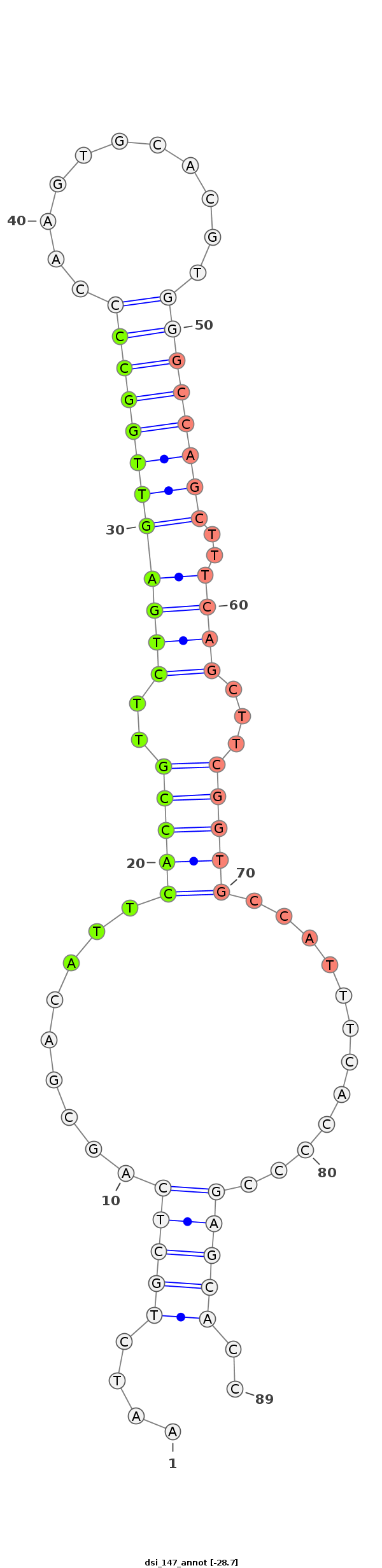
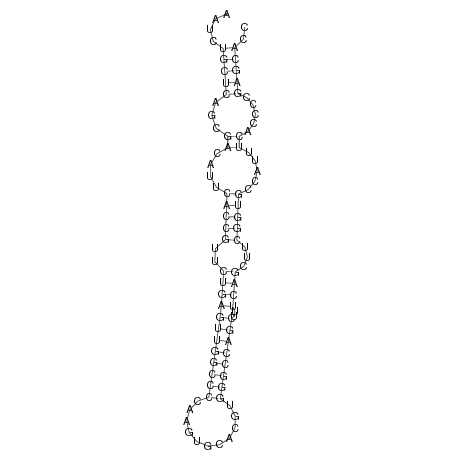| ..................................................ATTCACCGTTCTGAGTTGGCC.......................................................................................... |
21 |
0 |
1 |
1440.00 |
1440 |
1392 |
35 |
1 |
6 |
0 |
2 |
0 |
2 |
0 |
1 |
0 |
1 |
| ..................................................ATTCACCGTTCTGAGTTGGC........................................................................................... |
20 |
0 |
1 |
797.00 |
797 |
774 |
16 |
2 |
3 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCC......................................................................................... |
22 |
0 |
1 |
346.00 |
346 |
282 |
5 |
5 |
27 |
12 |
4 |
4 |
2 |
4 |
1 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGAC.......................................................................................... |
21 |
1 |
1 |
38.00 |
38 |
38 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGG............................................................................................ |
19 |
0 |
1 |
28.00 |
28 |
18 |
0 |
9 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTG............................................................................................. |
18 |
0 |
1 |
25.00 |
25 |
1 |
0 |
21 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
2 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGAC........................................................................................... |
20 |
1 |
1 |
15.00 |
15 |
15 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TTTCACCGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
14.00 |
14 |
13 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCT......................................................................................... |
22 |
1 |
1 |
14.00 |
14 |
14 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCT.......................................................................................... |
21 |
1 |
1 |
14.00 |
14 |
13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCCT........................................................................................ |
23 |
1 |
1 |
9.00 |
9 |
8 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGGTGGCC.......................................................................................... |
21 |
1 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TTTCACCGTTCTGAGTTGGC........................................................................................... |
20 |
1 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCCC........................................................................................ |
23 |
0 |
1 |
5.00 |
5 |
1 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGGTGCCAT.................................................... |
24 |
0 |
1 |
4.00 |
4 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCTTTCTGAGTTGGC........................................................................................... |
20 |
1 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGATTTGGC........................................................................................... |
20 |
1 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTTAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCGCCGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................TTCACCGTTCTGAGTTGGCCC......................................................................................... |
21 |
0 |
1 |
3.00 |
3 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...................................................TTCACCGTTCTGAGTTGGCC.......................................................................................... |
20 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGGTGCCATTTC................................................. |
27 |
0 |
1 |
3.00 |
3 |
0 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGCTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................GCACCAAGAGCATCAGAC........................ |
18 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGA........................................................................................... |
20 |
1 |
1 |
3.00 |
3 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACTGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCA......................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGCCCC......................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCTT........................................................................................ |
23 |
2 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................TCACCGTTCTGAGTTGGCCC......................................................................................... |
20 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGG........................................................................................... |
20 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGGTGGC........................................................................................... |
20 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCCCG........................................................................................ |
23 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................GACATTCACCGTTCTGAGTTGGCCC......................................................................................... |
25 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCCTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ACTCACCGTTCTGAGTTGGC........................................................................................... |
20 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................GCACCAAGAGCATCAGACTATAGAATT............... |
27 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTTGCCC......................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGG.......................................................... |
18 |
0 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGGGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................TCTGAGTTGGCCCCAAGTG................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCATCGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGCTCTGAGTTGGAC.......................................................................................... |
21 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................ACCGTTCTGAGTTGGCCC......................................................................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................CACCGCTCTGAGTTGGCCC......................................................................................... |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTCCTGAGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGGTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATCCACCGTTCTGAGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................ACCAAGAGCATCAGACTATAGAAT................ |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGATGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................TTCACCGTTCTGAGTTGAC........................................................................................... |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGCGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................GCACCAAGAGCATCAGACTATAGAA................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGCTCTGAGTTTGCCC......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................AGACTATAGAATTTACCATAAATA.... |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTATGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGCTT......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATCCACCGTTCTGAGTTGGC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTTACCGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTAGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGCTGGC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................AGACTATAGAATTTACCA.......... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGGTGCCA..................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGGGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAATTGGC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGCC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTAAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTGACCGTTCTGAGTTGGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGACC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................TCACCGTTCTGAGTTGGCC.......................................................................................... |
19 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGGTGGCCT......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTAAGTTGGC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................CTTCACCGTTCTGAGTTGGC........................................................................................... |
20 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGTGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTAGCC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTTGGCC......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................GCACCAAGAGCATCAGACTA...................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAATTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCTGAGTTGGTC.......................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................TTTCACCGTTCTGAGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................ATTCACCGTTCCGAGTTGGCCC......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................ATTTCACCGTTCTGAGTTGGCC.......................................................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGGTGCCATT................................................... |
25 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................GCCAGCTTTCAGCTTCGGT......................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................CATTCACCGTTCTGATTTGGCC.......................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................CATTCACCGTCCTGA................................................................................................. |
15 |
1 |
3 |
0.67 |
2 |
0 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................GACCGTTCGGAGTTGGCCCT........................................................................................ |
20 |
3 |
4 |
0.50 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................GACCGTTCGGAGTTGGCC.......................................................................................... |
18 |
2 |
5 |
0.40 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................GAGCATCAGACTGTA.................... |
15 |
1 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................TCACCCGGAGCACCA.................................... |
15 |
1 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ....................................ATCGGCTCAGCGAAATT............................................................................................................ |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |