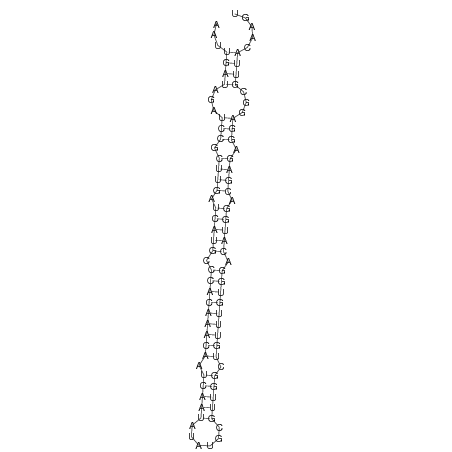
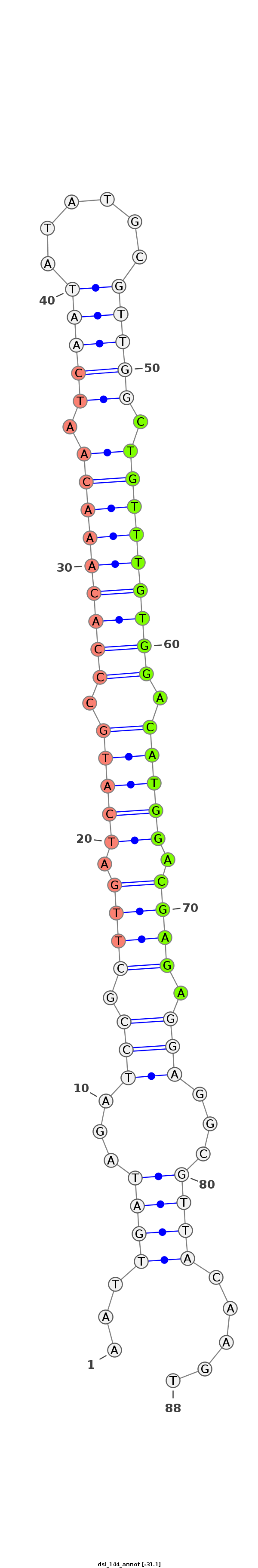
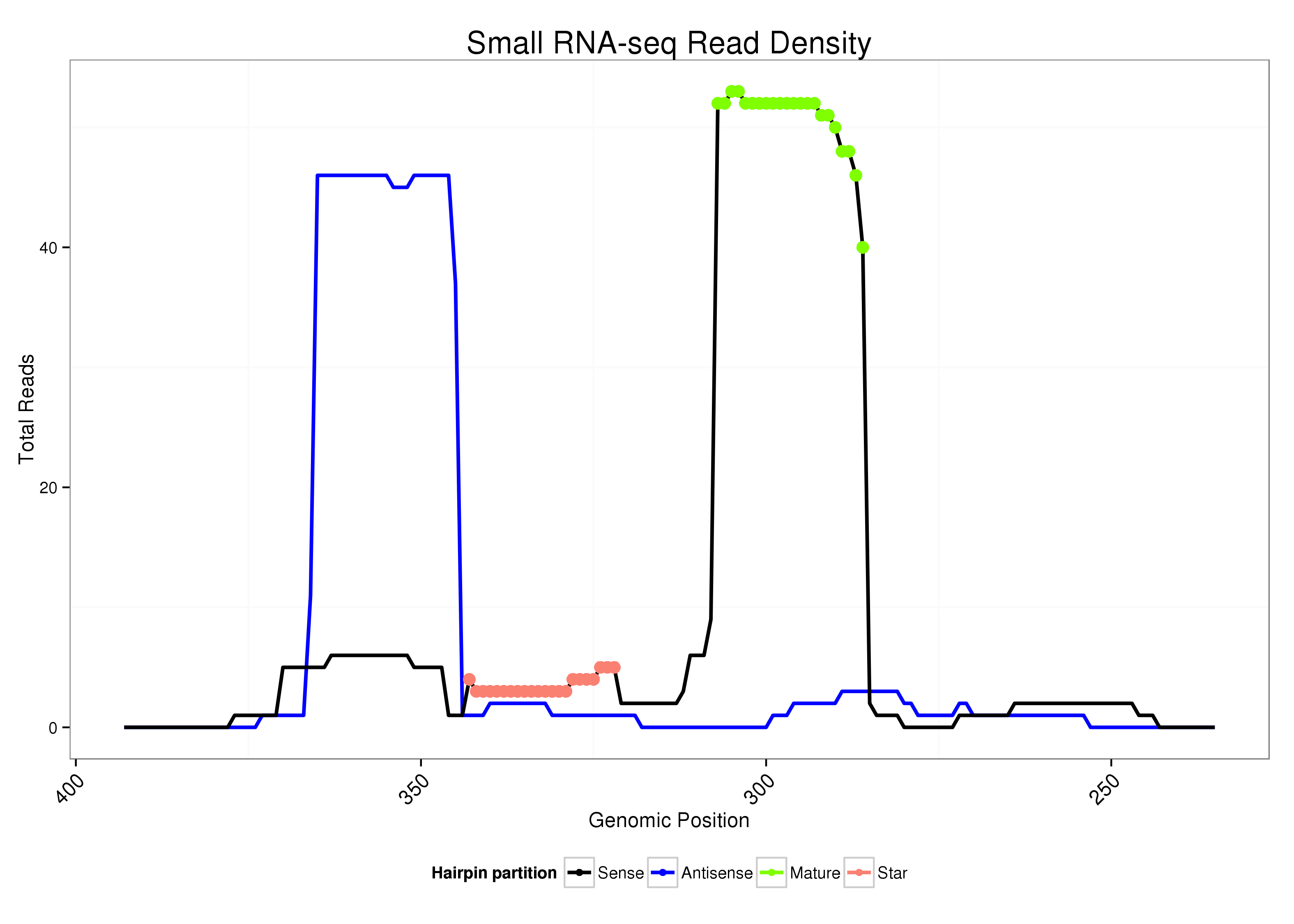
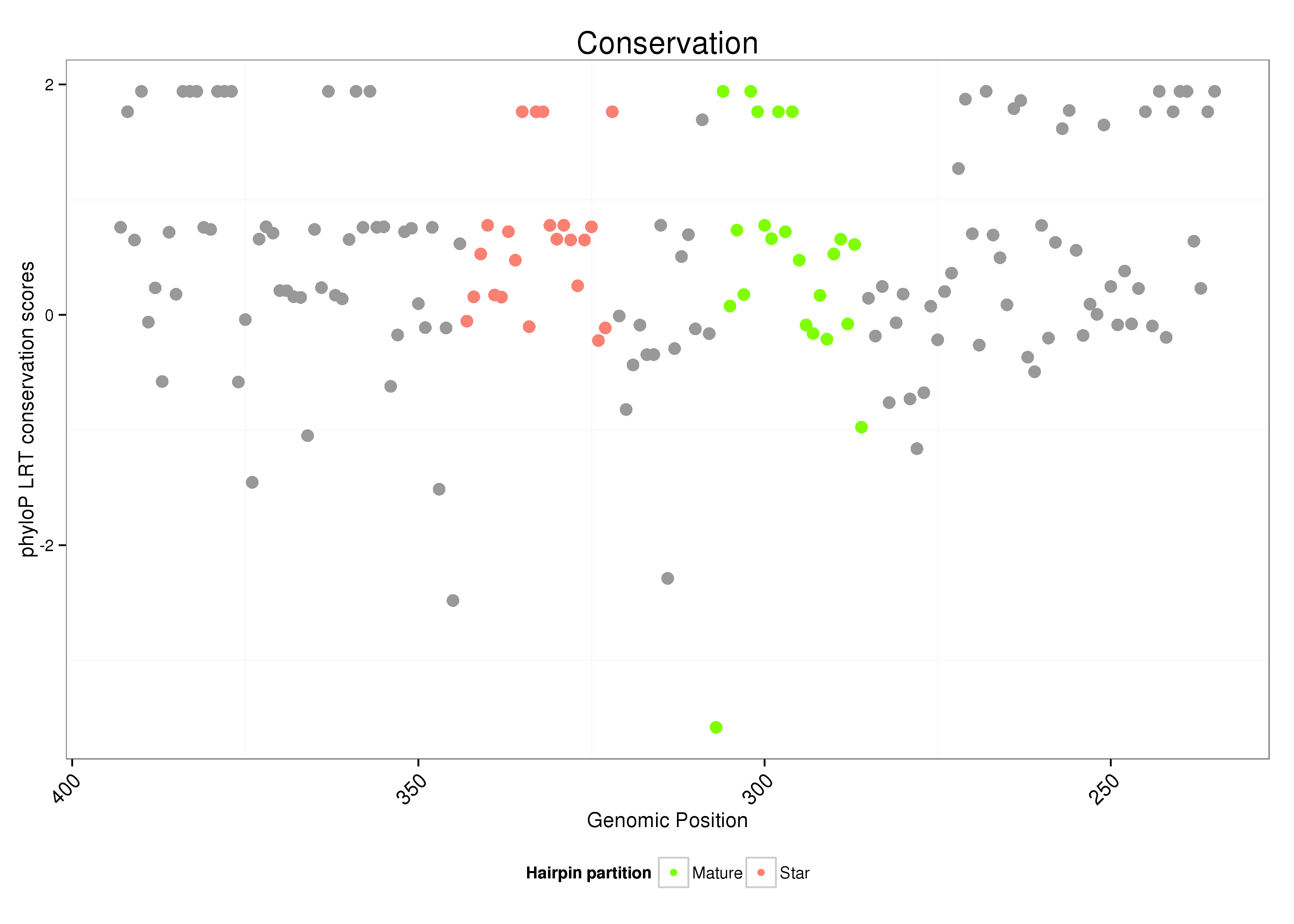
ID:dsi_144 |
Coordinate:chr3R_Mrandom_124:285-343 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
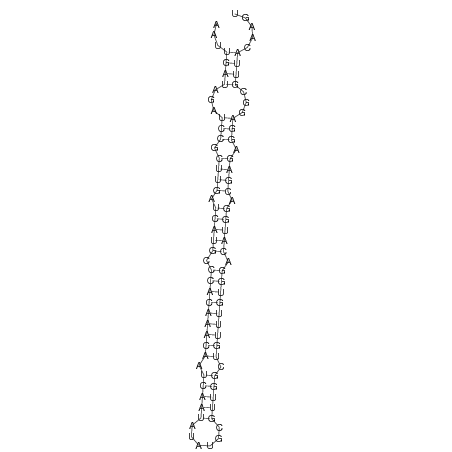 |
 |
 |
 |
| -30.3 | -30.3 | -30.1 |
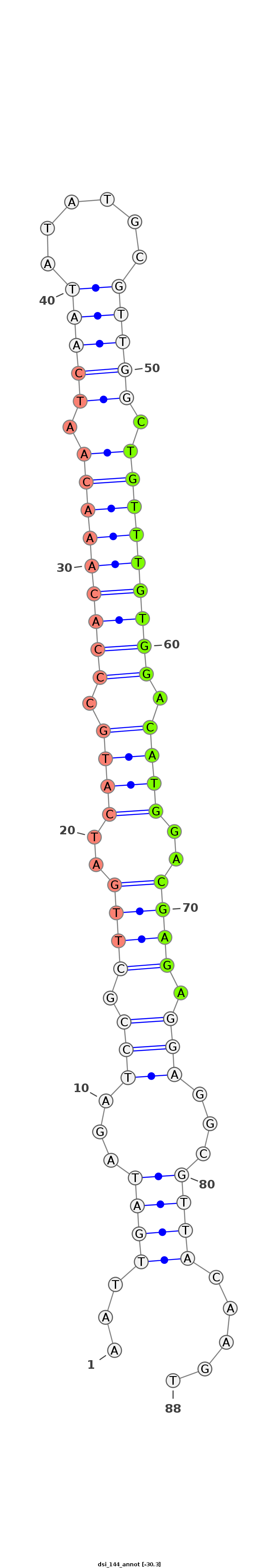 |
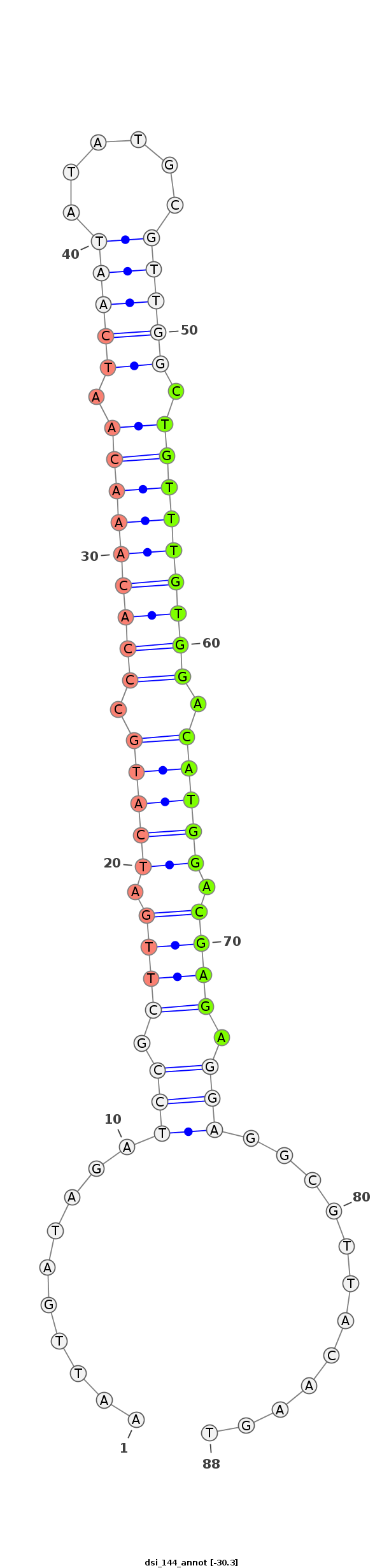 |
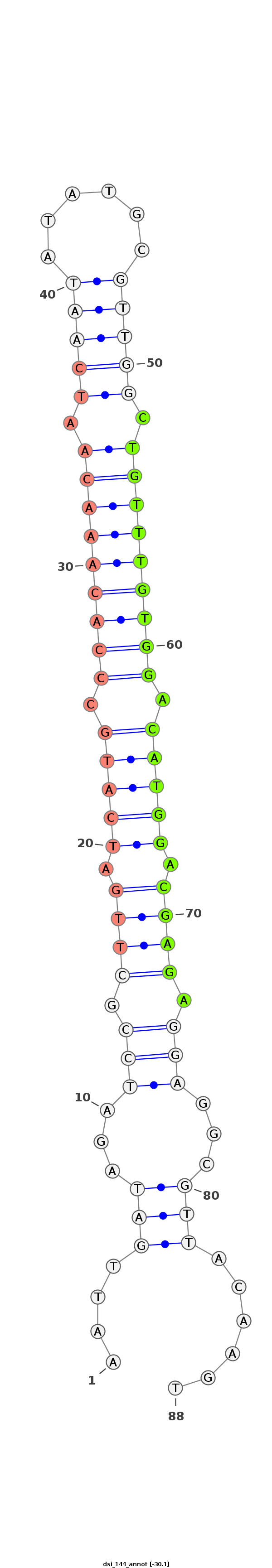 |
intergenic
No Repeatable elements found
|
AGCAATATGTTTATTTTGAGGAATAGCGTAATGGAAATTGATAGATCCGCTTGATCATGCCCACAAACAATCAATATATGCGTTGGCTGTTTGTGGACATGGACGAGAGGAGGCGTTACAAGTACAACATACGACACACACCGTGCATCAAACAACCGA
***********************************...((((...(((.((((.(((((.(((((((((.(((((......))))).))))))))).))))).)))).)))...)))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
SRR553486 Ovary |
M024 male body |
SRR553485 Ovary |
SRR553487 Ovary |
O001 Testis |
O002 Head |
SRR618934 Ovary |
M053 Female-body |
SRR553488 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................CTGTTTGTGGACATGGACGAGA................................................... | 22 | 0 | 1 | 38.00 | 38 | 13 | 11 | 4 | 6 | 0 | 2 | 0 | 0 | 0 | 1 | 1 |
| ......................................................................................CTGTTTGTGGACATGGACGAGG................................................... | 22 | 1 | 1 | 7.00 | 7 | 2 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CTGTTTGTGGACATGGACGAG.................................................... | 21 | 0 | 1 | 5.00 | 5 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................TTGATCATGCCCACAAACAATC....................................................................................... | 22 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| .......................TAGCGTAATGGAAATTGATAGATC................................................................................................................ | 24 | 0 | 1 | 3.00 | 3 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................GCTGTTTGTGGACATGGACGA..................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................TGGAAATTGATAGATCCGCTCGATCGT..................................................................................................... | 27 | 2 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CTGTTTGTGGACATGGACGAGAT.................................................. | 23 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TTGGCTGTTTGTGGACATGGAC....................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................ATCAATATATGCGTTGGC........................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................CTGTTTGTGGACATGGACGAGAAA................................................. | 24 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................GTTTGTGGACATGGACGAGAG.................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................TACGACACACACCGTGCATCA......... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................AACAATCAATATATGCGTTGGCTGT..................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................GCTGTTTGTGGACATGGACGAG.................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CTGTTTGTTGACATGGACGAGAT.................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TGTTTGTGGACATGGACGAGAGGAGG.............................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................AATGGAAATTGATAGATCCGCTCGATCGT..................................................................................................... | 29 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTGTCGACATGCAAGAGAGGAGG.............................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................TTGGCTGTTTGTGGACATGGA........................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................TAGCGTAATGGAAATTGATAGATCCGCT............................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................ATGGAAATTGATAGATC................................................................................................................ | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................TTGGCTGTTTGTGGACATGGATGAGGG.................................................. | 27 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................GTTGGCTGTTTGTGGACATG.......................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................GAAATTGATAGATCCGCTTGACA....................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................TGAGGAATAGCGTAATGGAAATTGAT..................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................GTACAACATACGACACACACCGTGCA............ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................CTGTTTGTGGACATGGACGAGAA.................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................TGATAGATCCGCTCGATCGTGCCCACAAAC........................................................................................... | 30 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TTTGTGGACATGGACGAGAGT................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GTTTATGTTGAGGAATA...................................................................................................................................... | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................GAAATTGATAGATCCGTTTGACA....................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................TTGTGGACAGGGACG...................................................... | 15 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................ACACACCGCGCGTCAAACC..... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................GGACGATAGGAGGCGT........................................... | 16 | 1 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ........................................GCAGATCCGCTTGATCA...................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| .GCAATATCTTTATTTGGAGTA......................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TCGGTTGTTTGATGCACGGTGTGTGTCGTATGTTGTACTTGTAACGCCTCCTCTCGTCCATGTCCACAAACAGCCAACGCATATATTGATTGTTTGTGGGCATGATCAAGCGGATCTATCAATTTCCATTACGCTATTCCTCAAAATAAACATATTGCT
************************************.....((((...(((.((((.(((((.(((((((((.(((((......))))).))))))))).))))).)))).)))...))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
O002 Head |
O001 Testis |
SRR553485 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................TATGTTGTACTTGTAACGCCT.............................................................................................................. | 21 | 0 | 1 | 35.00 | 35 | 32 | 3 | 0 | 0 | 0 | 0 |
| ...........................GTATGTTGTACTTGTAACGCC............................................................................................................... | 21 | 0 | 1 | 9.00 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| ............................TATGTTGTACTTGTAACGCCC.............................................................................................................. | 21 | 1 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ............................TATGTTGTACTTGTAACGCCG.............................................................................................................. | 21 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................GTATGTTGTACTTGTAACGCCT.............................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................AACGCCTCCTCTCGTCCATG................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................TTGTTTGTGGGCATGATCGAG................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TCGTCCATGTCCACAAACAGCC.................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGTGGGCATGATCAAGCGGAT............................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ATCAAGCGGATCTATCAAT.................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................GTGTGTCGTATGTTGTACT........................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................ATTTCCATTACGCTATTCC................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................GGGCATGATCAAGCGG.............................................. | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....TGTTTGATGCACGGTGTGTGG..................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................TATGTTGTACTTGTA.................................................................................................................... | 15 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................GTCGTATGGTGCATTTGTA.................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGTGGCCATGATTAAGAGG.............................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chr3R_Mrandom_124:235-393 - | dsi_144 | AGCAA-----TATGTTTAT--TTTGAGGAATAGCGTAATG-GAAATT-G--ATAGATCCGCTTGATCATGCCCACAAACAATCAAT--ATATG--------CGTTGGCTGTTTGTGGACATGGACGAGAGGAGGCG--TTAC----A-------AGTA------CAACATACGA--CACACACCGTGC----------ATCAAACAACCGA |
| droSec2 | scaffold_0:12486090-12486248 + | dse_135 | AGCAA-----TATGTTTAT--TTTGAGGAATAGCGTAATG-GAAATT-G--ATAGATCCGCTCGATCATGCCCACAAACAATCAGT--CTATG--------CGTTGGCTGTTTGTGGACATGGACGAGAGGAGGCG--TTAC----A-------AGTA------CAACATACGA--CACACACCGTGC----------ATCAACCAACCGA |
| dm3 | chr3R:9521062-9521220 - | AGCAA-----TATGTTTAT--TTTGAGGAATAGCGTAATG-GAAATT-G--ATAGATCCGCTCGACCATGCCCACAAACAATCAAT--CTATG--------GATTGGCTGTTTGTGGACATGGATGAGGGGTGGCG--TTAC----A-------AGTA------CAACATACGA--CACACACCGTGC----------ATCAACCAACCAA | |
| droEre2 | scaffold_4770:12114097-12114258 + | AGCAA-----TATTTTTAT--TTTGATGAATAGCGTAATG-GAAATT-A--ATAGATCCCCTCGACCATGCCCACAAACAATCAAT--CCATA--------GGTTGGCTGTTTGTGGACATAGACGAGAGGAGGCG--GTAC----GAGTACCAAGTA------CAACATAAAA------CACTGTGC----------ATCCACCAACCGA | |
| droYak3 | 3R:13829456-13829614 - | AGCAA-----TATGTTTAT--TTTGAGGAATAGAGTAATG-GAAATT-A--ATAGATTCCCTCGACCATGCCCACAAGCAATCAAC--CTATG--------GGTTGGATGTTTGTAGACATGGACGAGAGGAGCCG--GTAC----A-------AGTA------CAACATACGA--CACACCCTGTGC----------ATCCACCAACCGA | |
| droEug1 | scf7180000409802:453288-453444 + | AGCAA-----TATGTTTAT--TTTGACAAAAAGCTTAATG-GAAATT-A--ATAGATACACTCGACCATGCCCACACACAACCCAC--TA-TC-GTTTCTGGGTTGGTTGTGTGTGGACAAGGACGAGTAGTGACC--AGAC----A-------AGTA------CAACATAAGG--T--------CAC----------ATCCACCAACTGA | |
| droBia1 | scf7180000302402:2257639-2257797 - | AGCAA-----TATGTTTAT--TTTGAGGAAAAGCTTAATG-GAAATT-AGTATAGATTCCCCTGACCATGTCCGCACACAATCCAC--CTACT--AGTCTGC----GTTGTATGTGGACATGGACAGGAGGAGACG--AGAC----A-------AGTA------CAA----GTA--CACACCCTGTGC----------ATCCACCAAACGA | |
| droTak1 | scf7180000415380:348700-348870 - | AGCAA-----TATGTTTAT--TTTGACGAAAAGCTTAATG-GAAATT-A--TTAGATCCCCTCGACCATGCCCACACACAATCCCCCGTT-TC-GAATCTGGGTTGGATGTGTGTGGACATGGACGAGTGGAGATG--AGAGAC--A-------AGTA------CAACATAAGACACACACCCTGTGC----------AACCACCAACTGA | |
| droEle1 | scf7180000491047:1670847-1671018 + | AGCAT-----TATATTTAT--TTTTACGAAAAGCGTAACCCAAAATT-G--ATAGTTCCGCCCGACCATGCCCACACACATTCCTCAGCT-TC-GTGGCTGGGTTGGCTGTGTGTGGACATGAGCGAGCGGAGATG--GGACACATA-------AGTA------CAACATAAGA--CACACCCTGTGC----------ATCCACCAACTGA | |
| droRho1 | scf7180000769095:41483-41655 + | AGCAA-----GTTGTTTAT--TTTGACGAAAAGCGTAATA-GAAATT-A--ATAGTTCCACTCGACTATGCCCACACACATCCCCCACCA-TA-AAATCTGGGTCGGATGTGTGTGGACATGGGCGAGTGGATACACGGGACACATA-------AGTA------CAACATAAGA--CACACCCTGTGC----------ATCTATCAACTGA | |
| droFic1 | scf7180000453912:20388-20549 - | AGCAA-----TATGTTTAT--TTTTACGAAAAGCGTAACC-GAAATT-G--ATCATTTTACCCGGCCATGTCCATGCACAACCCAC--TCCTC-GAATC----TGGAATGTGTGCGGACATGGGCGAGTGAAAACG--GAGC----A-------AGTA------AAACATAACA--CACACCTTGTGC----------ATCCACCAACTGA | |
| droKik1 | scf7180000302706:531091-531275 - | AGCAGCAATATTTGTTTAT--TTTGTCGGAAAGCCTTACC-GAAATCTATTGTAGTTTCGCTCGACCATGCCCACACTTGGCCCAC--CTCTGAA-TAAGGCTTCGGCTAGGTGTGGACATGGGCGAGTGAAAACA--ACGC----GCA-----TGTTATATTTGAACGTAAGA--CACACCCTGTAC-------CTTATCCAGCAACTGA | |
| droAna3 | scaffold_13340:10521960-10522120 + | AGCAA-----TCTGTTTAA--TTTGACGAAAGTTTATACC-GAAATT-T--GCAGTTCTGTTCGATCCCGTCCACACACAGCCAAT--CTTTGGGTTCAAGGTTTGAGTCTGTGTGGGCGCAATCGCTTGGATACT--TTAG-----------------------ATACTAAGA--GACACTCCGTGT----------GACCATCAACTGA | |
| droBip1 | scf7180000396708:5537324-5537484 + | AGCAA-----TCTGTTTAA--TTTAAAGAACCTCTATACC-GAAATT-T--GTAGCTCCGCTTGATTACGTCCACACACAGCCAAT--CTTTGGGTTCAAGGTTTGAATCTGTGTGGACGTGATCGGTCGGATACT--TTGG----A---------TA------CA----AAGA--GACACTTCGTGT----------TACCACCAACTGA | |
| dp5 | 2:10072318-10072498 - | GGTAT-----AATGTTTTTTTTTTTGGGACAAACATAACC-GATACT----TTACCAATGCTCCACCACGCCCACACACATCCATA--CT-TA-ACCTATAGTCTGCCTGTGTGTGGACGATGCGGAGATCGCGCA--AGGA----AGGTACC-T-TA------TAACATAGAA--AGCATCCCGCATTTACCATCTACTCTACCAACTGA | |
| droPer2 | scaffold_0:10409010-10409190 + | GGTAT-----AGCGTTTTTTTTTTTTGGACAAACATAACC-GATACT----TTACCAATACTCCACCACGCCCACACACATCCATA--CT-TA-ACTATAAGTCTGCCTGTGTGTGGACGAGGCGGAGATCGCGCA--AGGA----AGGTACC-T-TA------TAACATAGAA--AGCATCCCGCATTTACCATCTACTCTACCAACTGA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 12/06/2013 at 02:32 PM