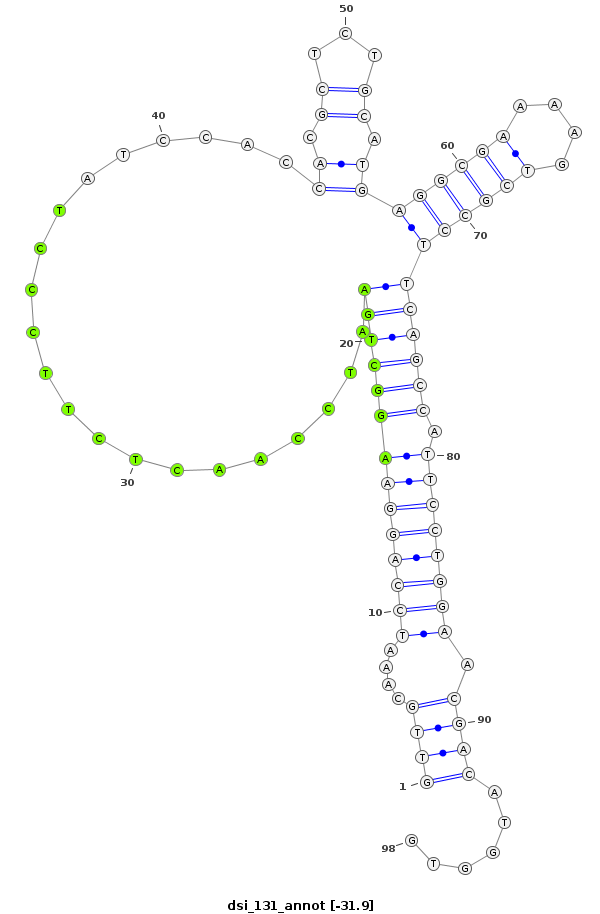
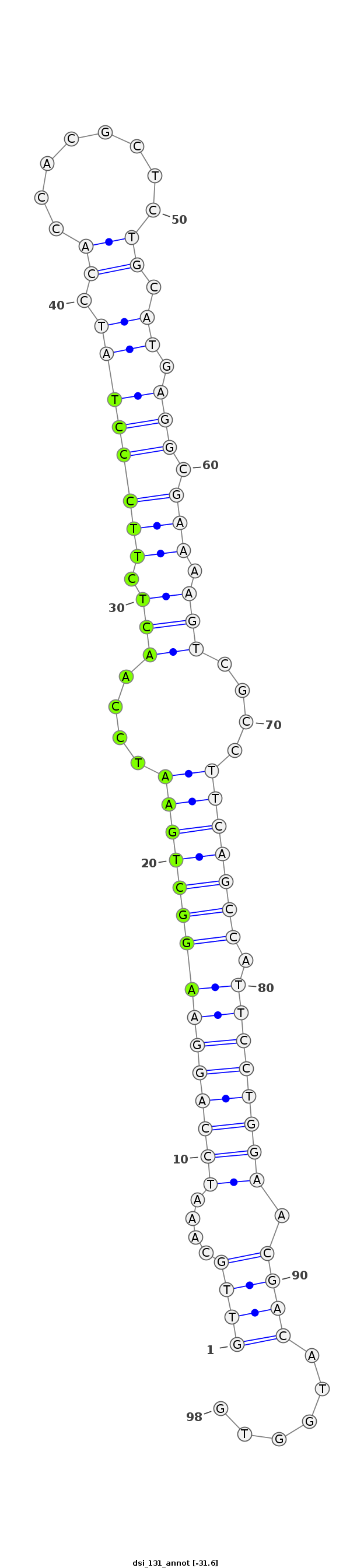
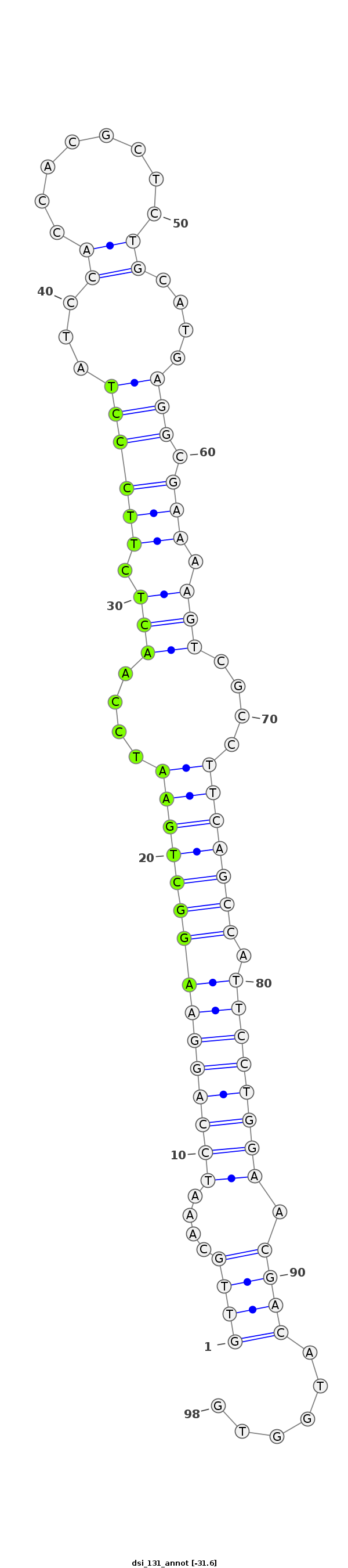
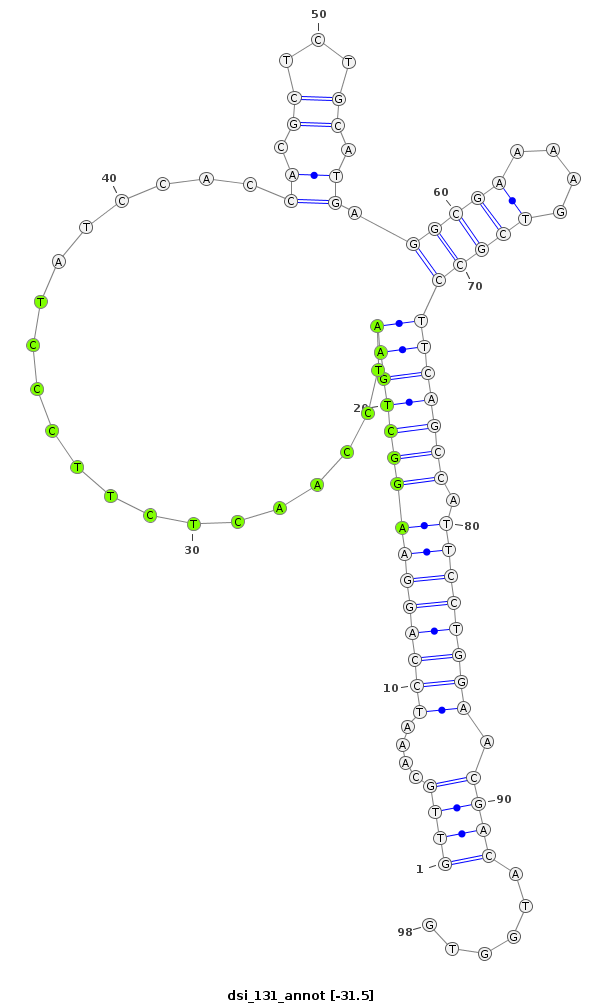
ID:dsi_131 |
Coordinate:3L:11184573-11184640 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
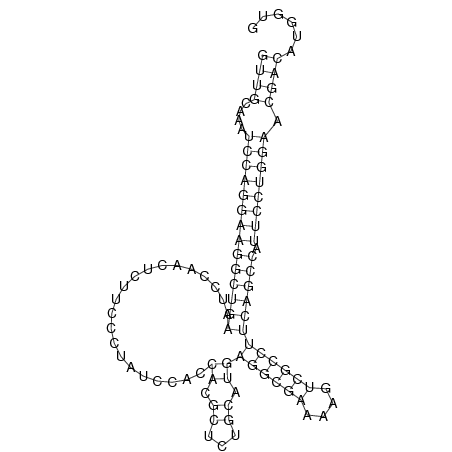 |
 |
 |
 |
| -31.6 | -31.6 | -31.5 |
 |
 |
 |
intergenic
No Repeatable elements found
|
ATTTGAGCTCCTAGTCGAGCTCAGTAAATATTGGTGTTGCAAATCCAGGAAGGCTGAATCCAACTCTTCCCTATCCACCACGCTCTGCATGAGGCGAAAAGTCGCCTTCAGCCATTCCTGGAACGACATGGTGGGGCTGATAGGGTCTGTGTCCACTGGCGGGGATTC
***********************************((((....((((((((((((((.....................((.((...)).))((((((....)))))))))))).)))))))).))))......*********************************** |
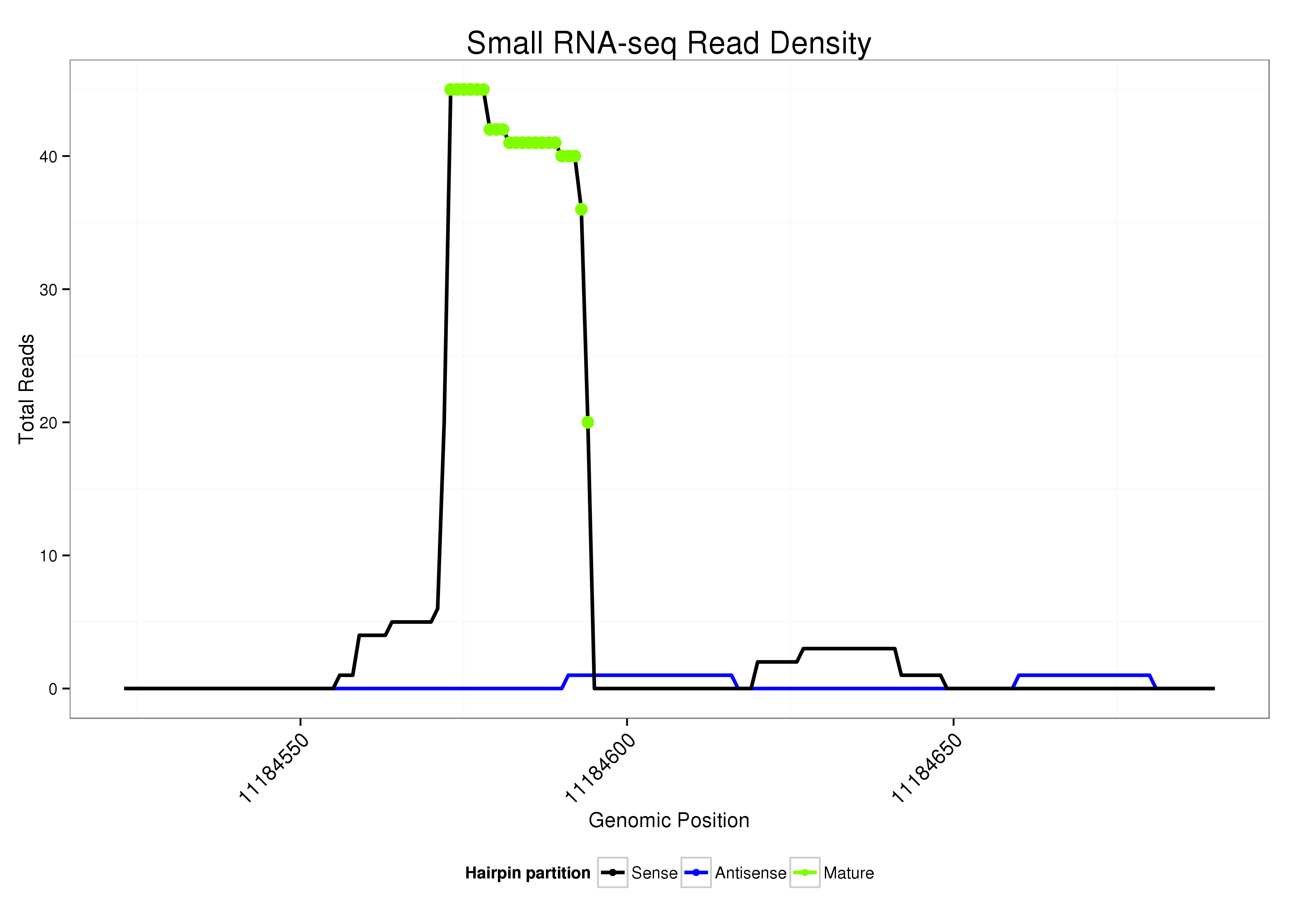
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O002 Head |
SRR553486 Ovary |
M025 embryo |
SRR553487 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................AGGCTGAATCCAACTCTTCCCT................................................................................................ | 22 | 0 | 1 | 20.00 | 20 | 19 | 0 | 0 | 0 | 1 | 0 |
| .................................................AAGGCTGAATCCAACTCTTCCC................................................................................................. | 22 | 0 | 1 | 11.00 | 11 | 10 | 0 | 0 | 1 | 0 | 0 |
| ..................................................AGGCTGAATCCAACTCTTCCC................................................................................................. | 21 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ....................................TTGCAAATCCAGGAAGGCTG................................................................................................................ | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .................................................AAGGCTGAATCCAACTCTTCC.................................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGGCTGAATCCAACTCTTCCCC................................................................................................ | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................AAAGTCGCCTTCAGCCATTCCT................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................................CCTTCAGCCATTCCTGGAACGA.......................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................GTGTTGCAAATCCAGGAAGGCTGAAT............................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................AATCCAGGAAGGCTGAATCCAACTCT..................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................AAACATTGGTGTTGTAAATC........................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GAAGGCTGAATCCAACTCTTCC.................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................AACATTGGTGTTGTAAATC........................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGGCTGAATCCAACTCTTCCCG................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGGCTGAATCCAACCCTTCCCT................................................................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................CAGCCATTCCTTGAAAGA.......................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TTCCTGGAACGACAT....................................... | 15 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................ACATTGGTGTTGTAAATC........................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................AAACATTGGTCTTGTAAATC........................................................................................................................... | 20 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................TCCACTGGCGTGGGTTC | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................TGTGTGCATTGGCGGGAATT. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
GAATCCCCGCCAGTGGACACAGACCCTATCAGCCCCACCATGTCGTTCCAGGAATGGCTGAAGGCGACTTTTCGCCTCATGCAGAGCGTGGTGGATAGGGAAGAGTTGGATTCAGCCTTCCTGGATTTGCAACACCAATATTTACTGAGCTCGACTAGGAGCTCAAAT
***********************************......((((.((((((((.((((((((((((....))))))((.((...)).)).....................))))))))))))))....))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|
| ....................................................................TTTTCGCCTCATGCAGAGCGTGGTGG.......................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| .........................................................................................................................................ATATTTACTGAGCTCGACTAG.......... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| .............................................................................................................................TCTGGACCACCAATATTTACT...................... | 21 | 3 | 6 | 0.50 | 3 | 0 | 0 |
| ..................................................CGAGTGGCTGAAGGCGAC.................................................................................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 0 |
| ...........................................................................................TGGAAATGGAAGAGTTGGATC........................................................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 |
| .................................................................................................................AGCCTTCCTGGCTTTGA...................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 |
| ..........................................................................................GTGTATAGGGAAGAATT............................................................. | 17 | 2 | 14 | 0.07 | 1 | 1 | 0 |
| ...........................................CGTCCCGGGAATGGCTGGA.......................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 1 | 0 |
| .................................................CCGAGTGGCTGAAGGCGAC.................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 3L:11184523-11184690 + | dsi_131 | ATTTGAGCT----CCTAGTCGAGCTCAGTAAATATTGGTGTTGCAAATCCAGGAAGGCTGAATCCAACTCTTCCCTATCCACCACGCTCTGCATGAGGCGAAAAGTCGCCTTCAGCCATTCCTGGAACGACATGGTGGGGCTGATAGGGTCTGTGTCCACTGGC-GGGGATTC |
| droSec2 | scaffold_0:4004472-4004615 + | ATTTGAGCT----CCTATCCGAGC------------------------TCAGGAAGGATGAATCCAACTCTTCCCTATCCACCACGCTCTGCATGAGGCGAAAAGTCGCCTTCAGCCATTCCTGGAACGACATAGTGGGGCTGATAGGGTCTGTGTCCACTGGC-GGGGATTC | |
| dm3 | chr3L:11819064-11819226 + | TTTTGCGCTTTTTCCT----AATCTCAGTATATATTGTTGTTCCAAACCCAGCAAGGCCGAATCCAACTCTTTCTTATACACCGTGCTCTGCAAGAAGCGAAAAGTCGCCCTCAGGCATTCCTGGAACGACGTAGCGGGGCTTATATGGT------CCACTGGCAGGGGATTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
Generated: 12/06/2013 at 02:32 PM