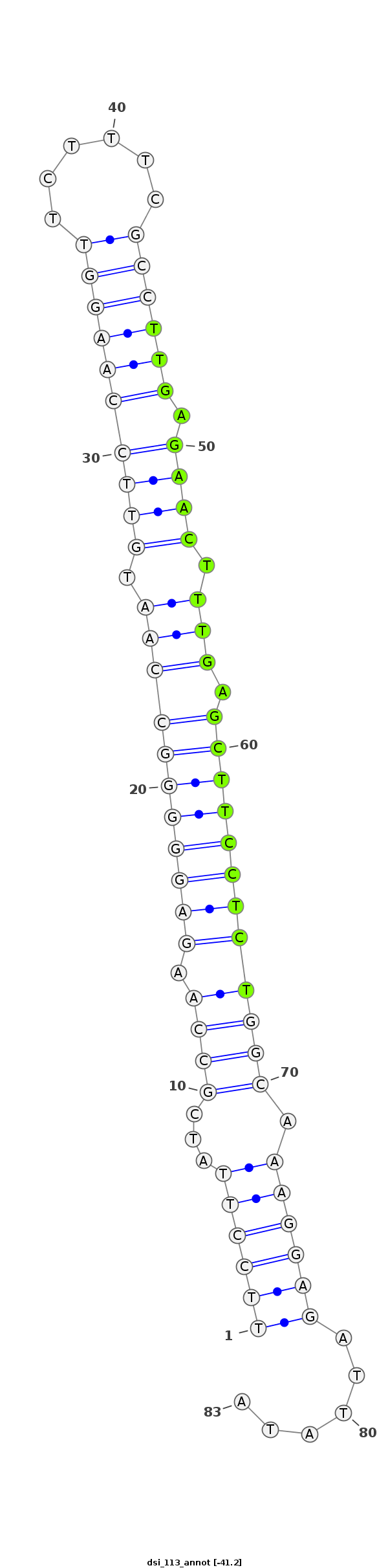
| ................................................................................TTGAGAACTTTGAGCTTCCTCT................................................... |
22 |
0 |
1 |
241.00 |
241 |
158 |
67 |
12 |
0 |
1 |
2 |
0 |
1 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTG.................................................. |
23 |
0 |
1 |
33.00 |
33 |
14 |
9 |
8 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCC................................................... |
22 |
1 |
1 |
31.00 |
31 |
22 |
8 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTC.................................................... |
21 |
0 |
1 |
28.00 |
28 |
21 |
4 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCT..................................................... |
20 |
0 |
1 |
23.00 |
23 |
16 |
6 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGG................................................. |
24 |
0 |
1 |
18.00 |
18 |
5 |
4 |
4 |
2 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTT.................................................. |
23 |
1 |
1 |
11.00 |
11 |
7 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGT................................................ |
25 |
1 |
1 |
7.00 |
7 |
2 |
0 |
3 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCG................................................... |
22 |
1 |
1 |
6.00 |
6 |
4 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................TTTCGCCTTGAGAACTTTGAGCTTCCT..................................................... |
27 |
0 |
1 |
5.00 |
5 |
0 |
0 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGG................................................ |
25 |
1 |
1 |
3.00 |
3 |
0 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGA................................................ |
25 |
1 |
1 |
3.00 |
3 |
0 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGC................................................ |
25 |
0 |
1 |
3.00 |
3 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGA................................................. |
24 |
1 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................TGAGAACTTTGAGCTTCCT..................................................... |
19 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCC..................................................... |
20 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................GCCTTGAGAACTTTGAGCTTCC...................................................... |
22 |
0 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGCT................................................ |
25 |
2 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCA................................................... |
22 |
1 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGTT............................................... |
26 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................CAAGGTTCTTTCGCCTTG...................................................................... |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCC...................................................... |
19 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................GTGAGAACTTTGAGCTTCCTCTGG................................................. |
24 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGATT.............................................. |
27 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................TCCTCTGGCAAAGGAGATTATAAT................................. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................CGCCTTGAGAACTTTGAGCTTCCT..................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................CTTGAGAACTTTGAGCTTCCTCT................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGACCTTCCTC.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCCG.................................................. |
23 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCCCT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCTT..................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................TTTGAGCTTCCTCTGGCAAA............................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................TGAGAACTTTGAGCTTCCG..................................................... |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................CTTTCGCCTTGAGAACTTTGAGCTTCCT..................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................ATAACTTTGAGCTTCCTCTG.................................................. |
20 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGGGAACTTTGAGCTTCCTCTG.................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................CGCCTTGAGAACTTTGAGCTTCCTCTG.................................................. |
27 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGGAA............................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................TGAGAACTTTGAGCTTCCTCTGG................................................. |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................TTGAGAACTTTGAGCTTCCTCTGAT................................................ |
25 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................CCTTGAGAACTTTGAGCTTCCTCTGG................................................. |
26 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................CGGCTTTGAGAACTTTGAGC.......................................................... |
20 |
3 |
16 |
0.13 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................TATGGACTAGAGGGGGCC............................................................................................... |
18 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................TGAGAACTTTGAACGTC....................................................... |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |