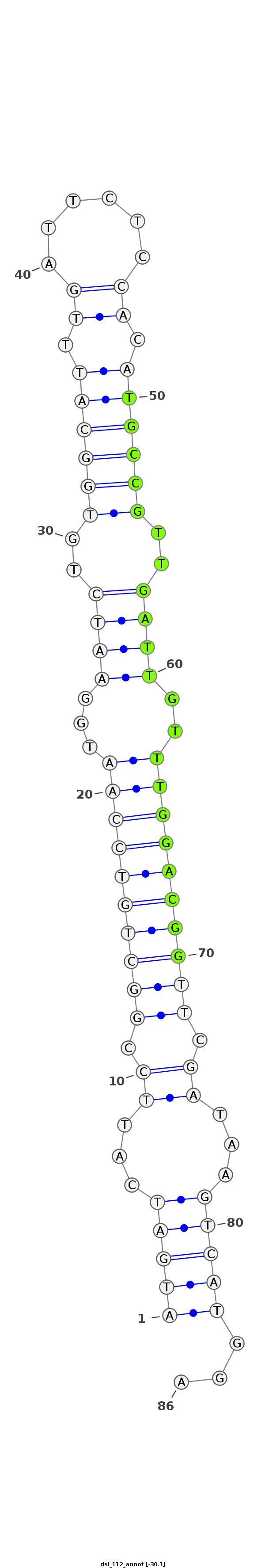
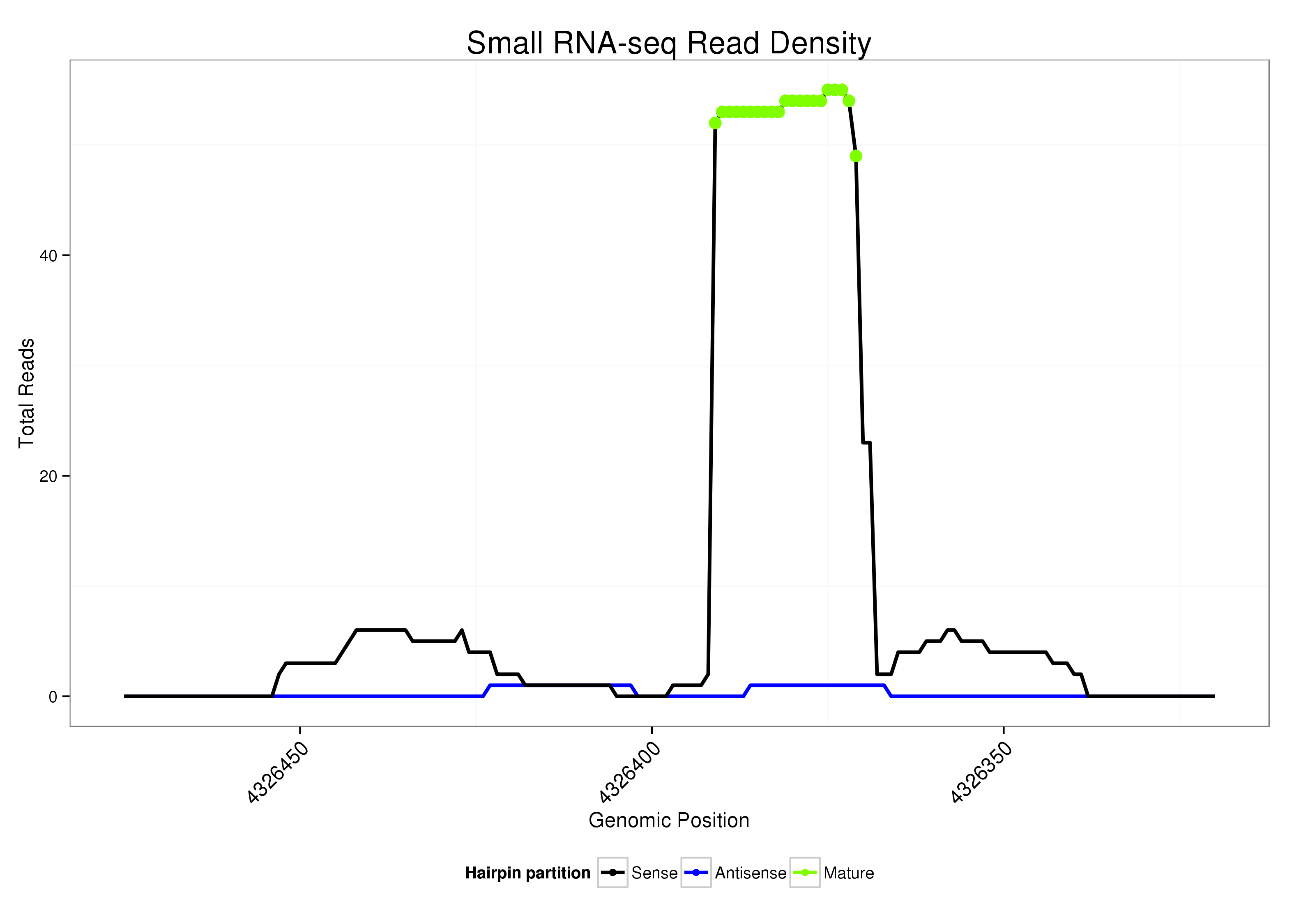
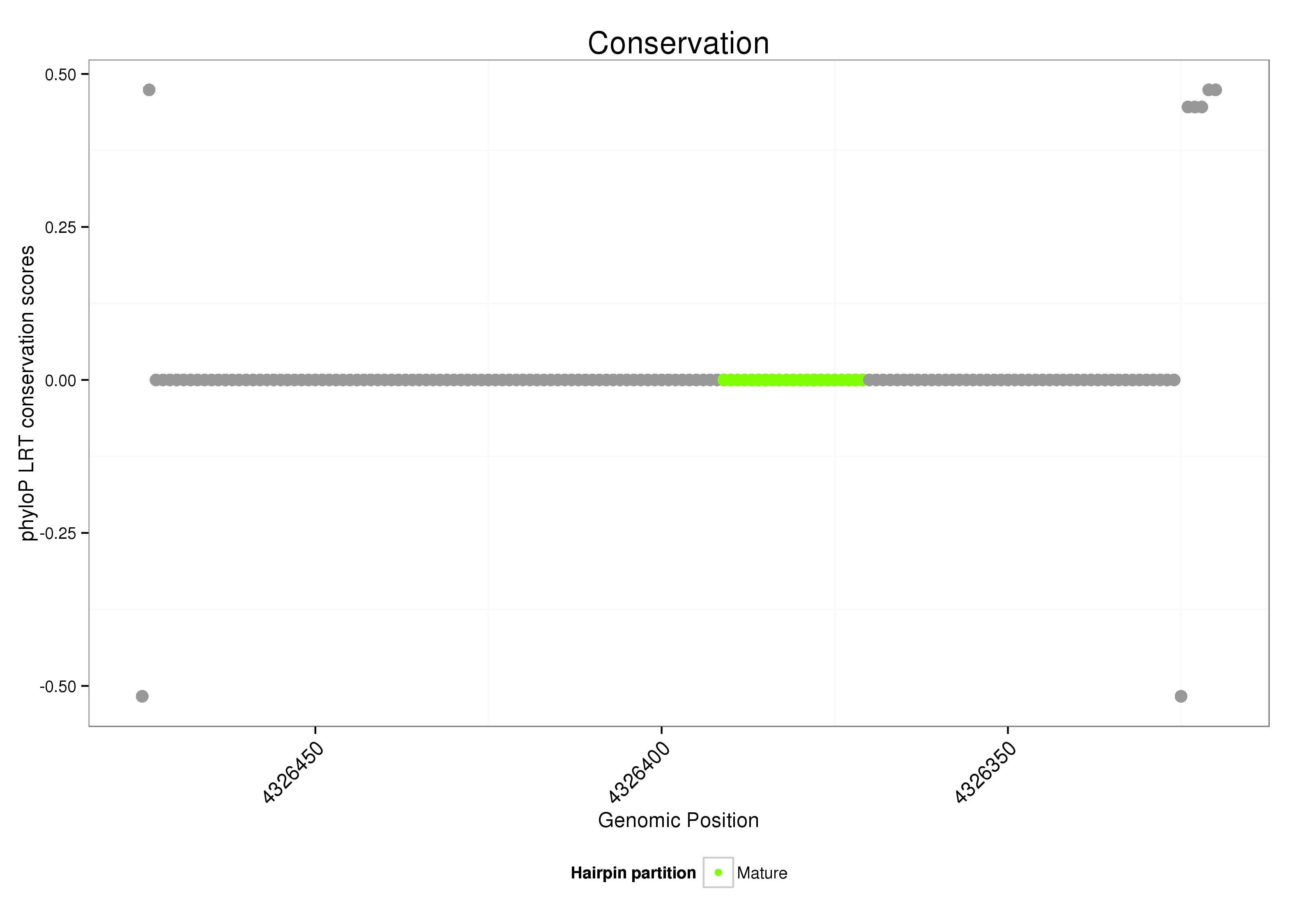
ID:dsi_112 |
Coordinate:3R:4326370-4326425 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
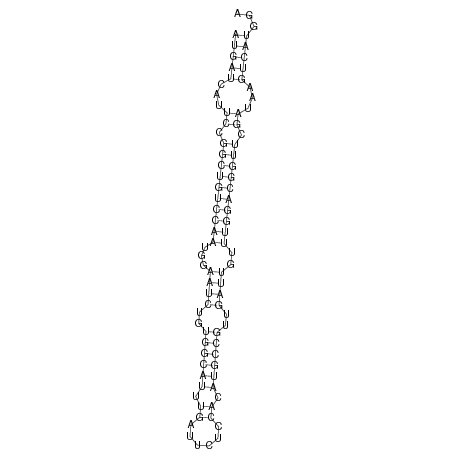 |
 |
 |
 |
| -29.2 | -29.1 | -29.1 |
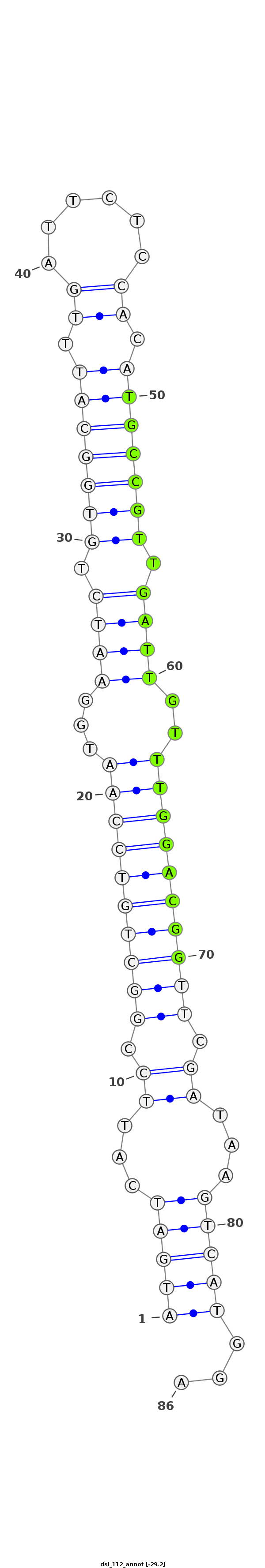 |
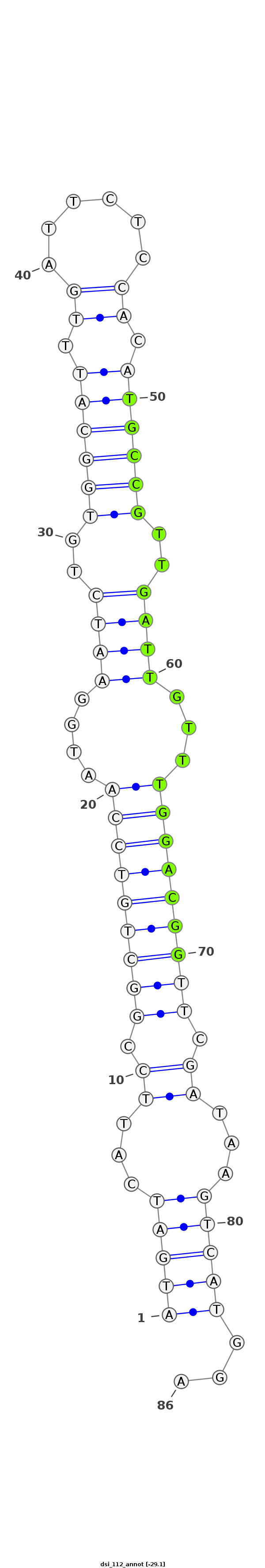 |
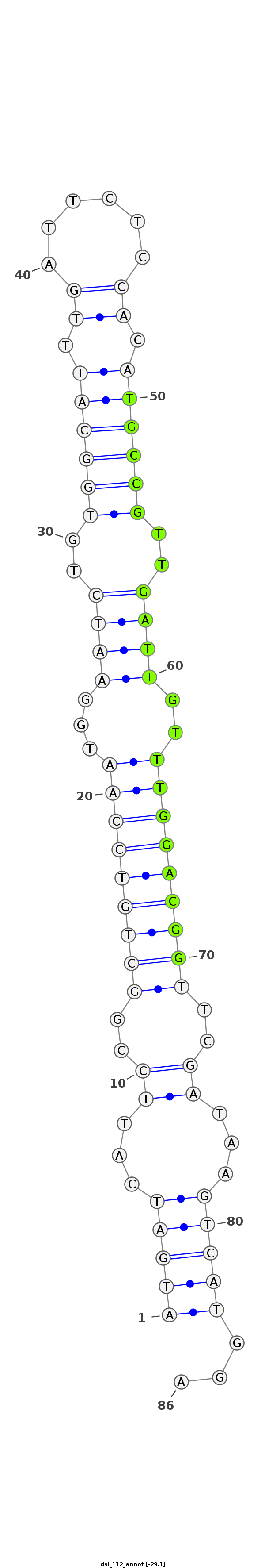 |
intergenic
No Repeatable elements found
|
CAGCTTAGCTCCACGGACGCCATAAGTATTGGCCCATGATCATTCCGGCTGTCCAATGGAATCTGTGGCATTTGATTCTCCACATGCCGTTGATTGTTTGGACGGTTCGATAAGTCATGGAGATCTTTATCGCTGTAGGCGATAAACGTACGGCAT
***********************************(((((...((.((((((((((...((((..((((((.((......)).))))))..))))..)))))))))).))...)))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
M025 embryo |
SRR553485 Ovary |
O001 Testis |
SRR553486 Ovary |
SRR553487 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TGCCGTTGATTGTTTGGACGG................................................... | 21 | 0 | 1 | 25.00 | 25 | 20 | 4 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................TGCCGTTGATTGTTTGGACGGTT................................................. | 23 | 0 | 1 | 20.00 | 20 | 15 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................TGCCGTTGATTGTTTGGACG.................................................... | 20 | 0 | 1 | 5.00 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TAAGTCATGGAGATCTTTATCGCTGTA................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................TAAGTATTGGCCCATGATCATTCCGGC........................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................TGTTTGGACGGTTCGATAAGTCATGGAGA................................. | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................GACGGTTCGATAAGTCATG..................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................TCATGGAGATCTTTATCGCTG..................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................CCATGATCATTCCGGCTGTCCAAT................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................ATGCCGTTGATTGTTTGGAC..................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................CGACTGTCCAATGGAATCTGTGGC....................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................TCCACATGCCGTTGATTGTTTGGACGGTT................................................. | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................CTGTCCAATGGAATCTGTGGCA...................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................TGGAGATCTTTATCG........................ | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................GCCGTTGATTGTTTGGACGG................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................TGCCGTTGATTGTTTGGACA.................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................GCCCATGATCATTCCGGCTGTC....................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................AAGTATTGGCCCATGATC................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................CGCCGTTGATTGTTTGGACGG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTCCAATGGAATCTGTGTCAT..................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CCCATGATCATTCCGGCTGTC....................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................TGCCGTCGATTGTTTGGAA..................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................CAGTGGATTGTTTGGACG.................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GCTCCAGGGACGCCA...................................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................GTTTCGACGGGTCGAT............................................. | 16 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................TTTGGGCGGTTGGATAA........................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ATGCCGTACGTTTATCGCCTACAGCGATAAAGATCTCCATGACTTATCGAACCGTCCAAACAATCAACGGCATGTGGAGAATCAAATGCCACAGATTCCATTGGACAGCCGGAATGATCATGGGCCAATACTTATGGCGTCCGTGGAGCTAAGCTG
***********************************...(((((...((.((((((((((..((((..((((((.((......)).))))))..))))...)))))))))).))...)))))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O002 Head |
|---|---|---|---|---|---|---|---|
| ........................................GACTAATCGAACCGTC.................................................................................................... | 16 | 1 | 2 | 3.50 | 7 | 0 | 0 |
| .........................................................................................CACAGATTCCATTGGACAGC............................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................GACTAATCGAACCGT..................................................................................................... | 15 | 1 | 4 | 1.00 | 4 | 0 | 0 |
| ....................................................CGTCCAAACAATCAACGGCAT................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 |
| ............................................TATGGAACCGTCCAAACGAAC........................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 |
| .........................................ACTAATCGAACCGTCTA.................................................................................................. | 17 | 2 | 20 | 0.25 | 5 | 0 | 0 |
| ........................................GACTAATCGAACCGTCTA.................................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 |
| ......................................................................................................GCAGAGCCGGAATGAACAT................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 |
| ..GCAGTATGTTTATCGCTTA....................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 3R:4326320-4326475 - | dsi_112 | CAGCTTAGCTCCACGGACGCCATAAGTATTGGCCCATGATCATTCCGGCTGTCCAATGGAATCTGTGGCATTTGATTCTCCACATGCCGTTGATTGTTTGGACGGTTCGATAAGTCATGGAGATCTTTATCGCTGTAGGCGATAA-------ACGTACGGCAT |
| droSec2 | scaffold_6:4347946-4348108 - | CAGCTAAGCTCCACGGACGCCATAAGTATTGGCCCATGATCATTCCGACTGTCCAATGGAATCTGTGGCATTTGATTCTCTGCATGACGTTGATTGTTTGGACGGTTCGATAAGTCATGGAGATCTTTATCGCTGTAGGCTATTACAGAGTGACGTACGGCAT | |
| dm3 | chr3R:17143181-17143330 + | AAGCTAAGCTCTACGAACGCCAAAACTATAGGCCTATGATCATTCCGACTGTCTAATGGAGTCTGTGGCACTTGATTCTCCACATGGCGTTGATTGTTTGGACGGTTCGATAAGTCGTGGAGATCTTTAGCGATGTA-----TTACAAAG--------TGCAT | |
| droEle1 | scf7180000490996:52187-52194 + | TA-----------------------------------------------------------------------------------------------------------------------------------------------------------TGGCAT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 12/06/2013 at 02:31 PM