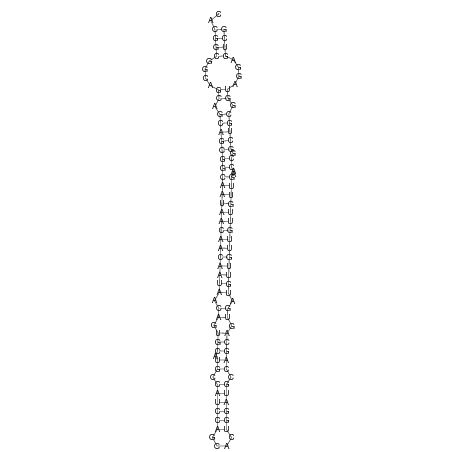

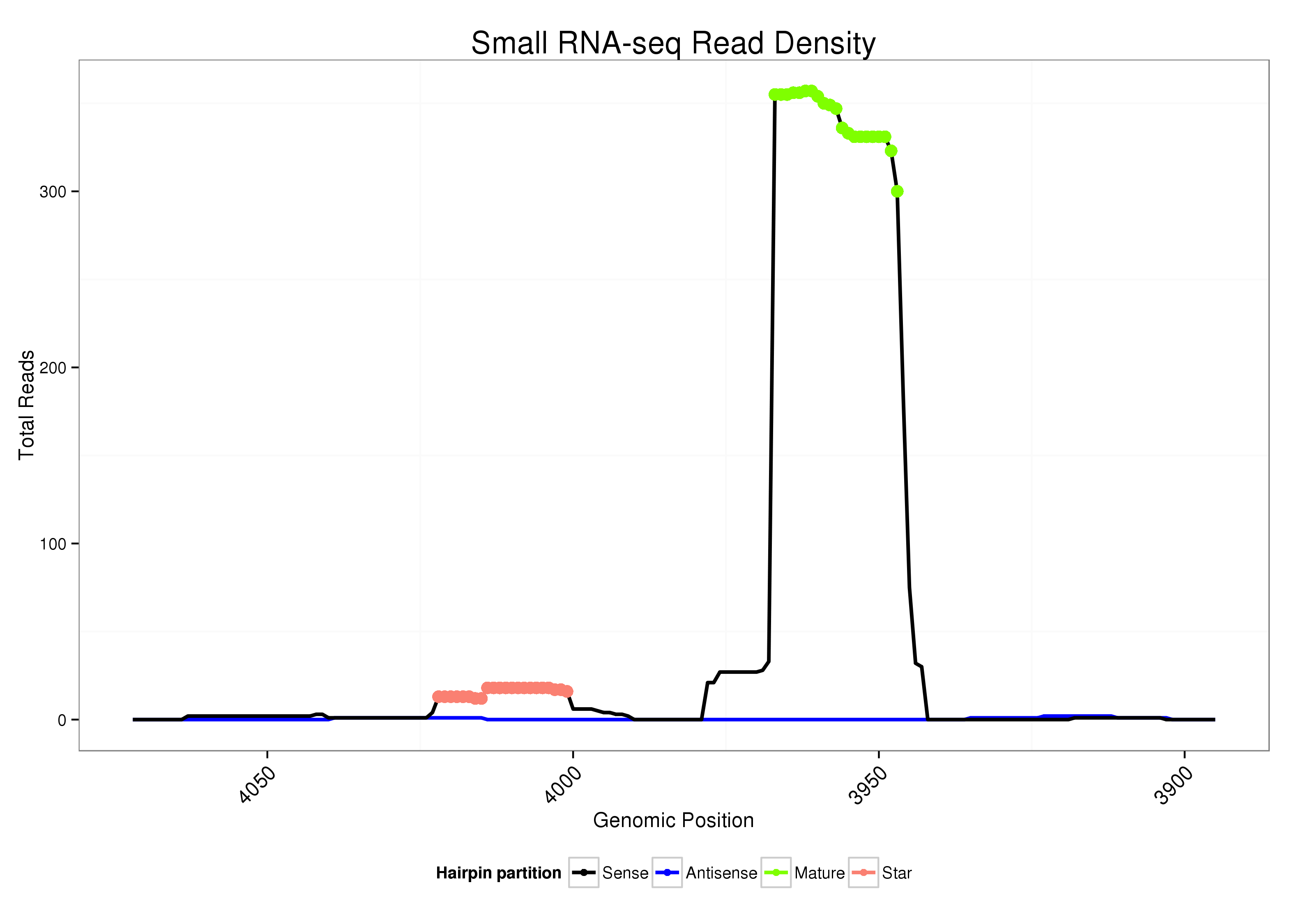
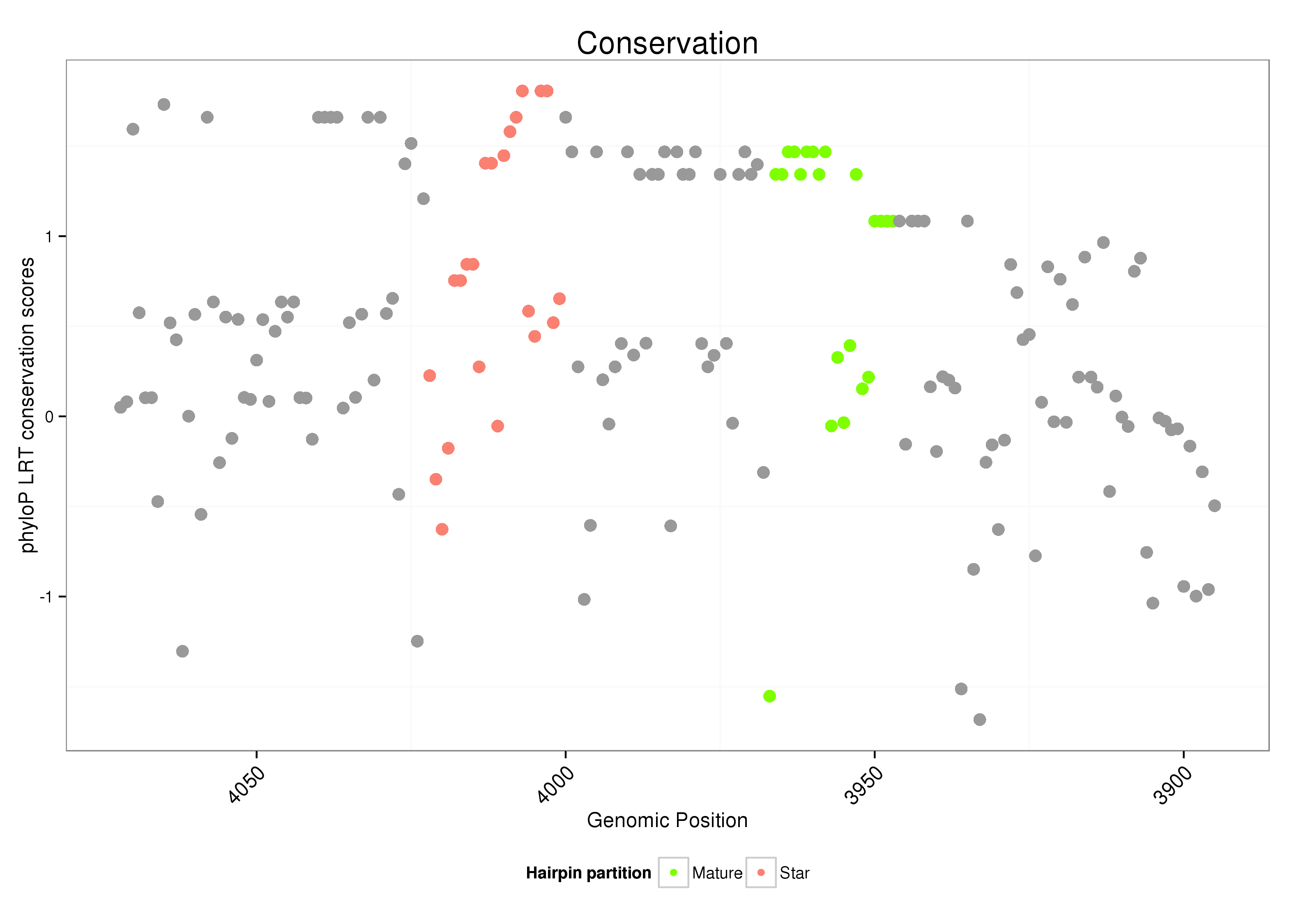
ID:dsi_108 |
Coordinate:chrX_Mrandom_244:3945-4022 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -48.7 | -47.8 | -47.8 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCGTTACTCCTCGACAGCAGTTCTCCTCTTCTCGCCACGGCGGCAGCAGCAGCGGCAATAACAACAATAACAGTGCATGCCATCCAGCACTGGATGCCAGCAGTGATGTTGTTGTTGTTGAACCGGCTGCGGTAGGAGTCGCACAGGAGAAAGAGAAAGAGCCGGGGCAGAAGCCAGA
***********************************..((((....((.(((((((((((((((((((((.((.(((.((.((((((....)))))).))))).)).))))))))))))))..)).))))).))....))))************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 Ovary |
SRR553485 Ovary |
SRR553486 Ovary |
SRR553488 Ovary |
M025 embryo |
M023 head |
O001 Testis |
SRR618934 Ovary |
M024 male body |
O002 Head |
GSM343915 embryo |
M053 Female-body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................ATGTTGTTGTTGTTGAACCGG.................................................... | 21 | 0 | 1 | 117.00 | 117 | 44 | 23 | 12 | 26 | 1 | 0 | 0 | 10 | 0 | 0 | 1 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGC................................................... | 22 | 0 | 1 | 106.00 | 106 | 22 | 24 | 13 | 7 | 16 | 15 | 0 | 5 | 4 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCT.................................................. | 23 | 0 | 1 | 38.00 | 38 | 6 | 0 | 4 | 4 | 13 | 8 | 0 | 2 | 1 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCTGC................................................ | 25 | 0 | 1 | 29.00 | 29 | 5 | 5 | 12 | 0 | 1 | 1 | 0 | 5 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCG..................................................... | 20 | 0 | 1 | 22.00 | 22 | 7 | 3 | 1 | 2 | 1 | 5 | 1 | 1 | 1 | 0 | 0 | 0 |
| ..................................................AGCGGCAATAACAACAATAACA.......................................................................................................... | 22 | 0 | 1 | 8.00 | 8 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 1 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACC...................................................... | 19 | 0 | 1 | 8.00 | 8 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 1 | 1 | 1 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGTTGTT.............................................................. | 22 | 0 | 1 | 8.00 | 8 | 0 | 0 | 0 | 0 | 2 | 0 | 4 | 0 | 0 | 1 | 0 | 1 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCC.................................................. | 23 | 1 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGTT................................................................. | 19 | 0 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGT.................................................................. | 18 | 0 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GATGTTGTTGTTGTTGAACCGGCT.................................................. | 24 | 0 | 1 | 4.00 | 4 | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCTA................................................. | 24 | 1 | 1 | 3.00 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................CCAGCAGTGATGTTGTTGTT.............................................................. | 20 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TAACAACAATAACAGTGCATGCCA................................................................................................ | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........CTCGACAGCAGTTCTCCTCTTCT.................................................................................................................................................. | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CAGCGGCAATAACAACAATAACA.......................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCTG................................................. | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCA.................................................. | 23 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGTTGT............................................................... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGTTGTTG............................................................. | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGA................................................... | 22 | 1 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................CCAGCAGTGATGTTGTTGTTGT............................................................ | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCTT................................................. | 24 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................AGCGGCAATAACAACAATA............................................................................................................. | 19 | 0 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................GATGTTGTTGTTGTTGAACCGGCC.................................................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TAACAACAATAACAGTGCA..................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................CTCGCCACGGCGGCAGCAGCAGCGGC.......................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................TAACAACAATAACAGTGCATGCC................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTGTTGTTGTTGAACCGGCTGC................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGG................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................GTTGTTGTTGAACCGG.................................................... | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGT................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................AGGTTGTTGTTGTTGAACCGGCT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................GTCTTCTCGCCACGGTGGAAG.................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................CCAGCAGTGATGTTGTTGTTG............................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCTGCTA................................................. | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TAACAACAATAACAGTGC...................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAATCGGC................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GAAAGAGCCGGGGCA......... | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCTAAA............................................... | 26 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGGCAAA................................................ | 25 | 3 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGGACCGGC................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................TAACAACAATAACAGTGCATG................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................ATGTTGTCGTTGTTGAACCGGC................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................TGTTGTTGAACCGGC................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CAGCGGCAATAACAACAATAAC........................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................TGCCAGCAGTGATGTTGTTG................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GATGTTGTTGTTGTTGAACCG..................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................ATGTTGTTGTTGTTGAACCGT.................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................TGATGTTGTTGTTGTTGAACCGGCT.................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................TGATGTCGTTGTTGTTGAA........................................................ | 19 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGCGGCAATAACAACAATAACAA......................................................................................................... | 23 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................ATGTTGTTGTTGAACCGGCA.................................................. | 20 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GCCACTAGTGATTTTGTTGTT.............................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................CAGCGGCAATAACAACAATAAAAA......................................................................................................... | 24 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................AGTTCTCCTCTTCCC................................................................................................................................................. | 15 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGAGTCGCACAGG............................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................GCCACTAGTGATTTTGTTGT............................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................TGATGTCGTTGTTGTTGAAA....................................................... | 20 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGTGATTTTGTTGTTGT............................................................ | 17 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................GCAGCAGCGGCCATATCA................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TCTGGCTTCTGCCCCGGCTCTTTCTCTTTCTCCTGTGCGACTCCTACCGCAGCCGGTTCAACAACAACAACATCACTGCTGGCATCCAGTGCTGGATGGCATGCACTGTTATTGTTGTTATTGCCGCTGCTGCTGCCGCCGTGGCGAGAAGAGGAGAACTGCTGTCGAGGAGTAACGG
*************************************((((....((.(((((.((..((((((((((((((.((.(((((.((((((....)))))).)).))).)).))))))))))))))))))))).))....))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M053 Female-body |
SRR553485 Ovary |
|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................GCCGTGGCGAGAAGAGGAGAACTG................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................TGTGCGACTCCTACCGCAGCCGGTT........................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| .....................................................................................................................................................AGAGGAGAACTGCTGTCGAGG........ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ........................TCTTTCTCCTGTGCGACTCCC..................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..................................................................................CATCCATTGCTGGAGGGCTT............................................................................ | 20 | 3 | 7 | 0.29 | 2 | 2 | 0 | 0 |
| .......................................................................................AGTCCTGGATCGCATGCA......................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 |
| .................................................................................................GCCATCCACTGTTAGTGTT.............................................................. | 19 | 3 | 16 | 0.13 | 2 | 0 | 0 | 0 |
| ......................................................................................................................TATGGCCGCTGCTGCT............................................ | 16 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 |
| ............................................TATCGCGGCCGGTTC....................................................................................................................... | 15 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 |
| ......................................................................................................................TATGGCCGCTGCTGAGGCCG........................................ | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TATGGCCGCTGCTGCCACCG........................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TATGGCCGCTGCTGCTG........................................... | 17 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| ..................................................................................................................TTGATATGGCCGCTGCTG.............................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| ....................................................................................................................GATATTGCCGCTGCTG.............................................. | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chrX_Mrandom_244:3895-4072 - | dsi_108 | CCGTTACTCCTCGACAGCA-------------GTTCTCCTCTTCTCGCCACGGCGGCAGCAGCAGCGGCAATAACAACAATAA---------------------CAGTGCATGCCATCCAGCACTGGATG---CCAGCA---GTGATG---TTGTTGTTGTTGAACCGGCTGCGGTAGGAGTCGCACAGGA---GAAAGAGAA---------AGAG---CCGGGGCAGAAGCCAGA |
| droSec2 | scaffold_4:2149964-2150135 + | dse_197 | CCGTTACTCCTCGACAGCA-------------GTTCTCCTCTTCTCGCCACGGCGGCAGCAGCAGCGGCAATAACAACAATAA---------------------CAGTGCATGCCATCCAGCACTGGATG---CCAGCA---GTGATG---TTGTTGTTGTTGAACCGGCTGCGGTAGGAGTCGCACAGGA---GA------A---------AGAG---CCGGAGCAGAAGCCAGA |
| dm3 | chrX:4579780-4579933 - | CCGTTACT-------------------------------------CGCCACGGCGGCAGCAGCAGCGGCAATAACAACAATAA---------------------CAGTGCATGCCATCCAGCACTGGATG---CCAGCA---GTGATG---TTGTTGTTGTTGAACCGGCAGCGGTAGGAGTCGCACAGGA---GGAAGAGGA---------AGAG---CCGGAGCAAAGGCCAGA | |
| droEre2 | scaffold_4690:1957110-1957273 - | CTGTTACTCCGCGACATCA-------------GTTGTCCTCTTCTCGCCACGGCGGCAGCAACAAT---AACAACAACAATAA---------------------CAGTGGGTGCCATCCAGCATTGGATG---TCAGCA---GTGGTG---TTGTTGTTGCTGCACCGGCAGCGATAGGAGGAGCACAG------GAAGAGGA---------AGAG---CCGGAGCAG-------- | |
| droYak3 | X:3860422-3860596 + | CCGTTACTTCGCGACAGCA-------------GTTGTCCTCTTCCCGCCACGGCGGTAGCAACAAT---------AACAATAATAACAGCAACAGCAACAACAGCAGTGGGTGCCATTCAGCATTGGATG---CCAACA---GTGGTG---TTGTTGTTGTTGCACCGGCAGCGGTAGGAGGAGCACAG------GAAGAGGA---------AGAG---CC------------AGA | |
| droEug1 | scf7180000409548:355299-355458 - | CCGTTAATCCCAGACAGCA-------------GCTGTCCTCTTCTCGCCCCAACGGCAACA---------------ACAATAA---------------------CAGTGATTGTCATCCAGCACTGGATG---CCAACA---GTACTG---TTGTTGTTGTTGCACCGGCAGCGGAAAGTGAGCCTGAG------GAGGA------------AGAACCGCCTGAAGAAGATGAAAA | |
| droBia1 | scf7180000301703:117349-117538 + | CCGTTAGTCCGCGACAGC-AGCAGCAACAGCAGTTGTCCTCTTCTCGCCACGGCGGCAGCAA---------CAATAACAATAA------------------CGACAGTGGGTGCCATCCAGCACTGGATG---CCAGCA---GTAGTGTTGTTGTTGTTGTTGCACCGGCAGCGGGAAGGGACGAGCA---ACTGGAAGAG------GCCGTGGAG---CTGGAAGTGGTGGTGAA | |
| droTak1 | scf7180000414746:63451-63602 - | GC----------AGCAGCA-------------GTTGTCCTCTTCTCGCCACGGCGGCGGCAGCAA------CAATAACAATAA---------------------CAGTGGGTGCCATCCAGCACTGGATG---CCAGCAGCAGTAGTGTTGTTGTTGTTGTGGCGCCGGCAGCGGGAAGAG---AACAGGAACTGG------AAGGGG----AG-------------------AGG | |
| droEle1 | scf7180000491011:1727310-1727462 + | CCGTTTCTCCGCGACAAC-AGC---AACAGCAGTTGTCCTCTTTTCGCCACGGCGGCAA------------CAATAACAATAA---------------------CGGTGAGTGCCATCCAGCATTGGATGGATCCGGCA---G---TG---TTGTTGTTGTTGCACCGGCAGCGGGAAAGGA---------ACTAG------AAGAGAC---AGAA---C---------------- | |
| droRho1 | scf7180000778083:144963-145121 - | CCGTCACTCCGCGACAAC-AGC---AACACCAGTTGTCCTCTTCTCGCCACGGCGGCAA------------CAATAACAATAA---------------------CAGTGAGTGTCATCCTGCATTGGATGGATCCAGCA---GTGTTG---TTGTTGTGGTTGCACCGGCGGCGGGAAGCGA---------ACTCGAAGGG------GC---AGAA---C-------------GGG | |
| droFic1 | scf7180000453775:107965-108123 + | CCGTTACTCCACGGCAAC-AGCAGAAACAGCATTTGTCCTCTTCTCGCCACGGCAGCAA------------CAATAACAATAA---------------------CAGTGAGTGCCATCCAGCACTGGATG---CCAGCA---GT---G---TTGTTGTTATTGCACCGGCAGCGGGGAGAGG---------TC---------A---------AA---CGTCGGATCAGGATGCAT- | |
| droBip1 | scf7180000396383:83283-83399 + | TTGTTAATCCACGCCAGCC------AATATC-GCCGGTATCTGCACGCCCTGGCAGCACCATCAG------------CAGCAA------------------CAGTAGTTGCTCCAGTCCAGCATTGGACT---CCAGCA---GTGTTG---TTGTTGTGGGTG------------------------------------------------------------------------- | |
| droGri2 | scaffold_15110:15667277-15667348 + | CCGCCGCTCATAGGCGGTG-------------GTG-TACCTCCTCCGCCACAGCAGCAACAGCTGCAGCAACAACAACAACAA---------------------CAG--------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 12/06/2013 at 02:31 PM