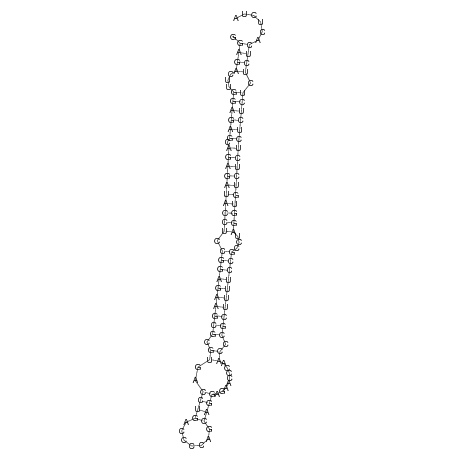
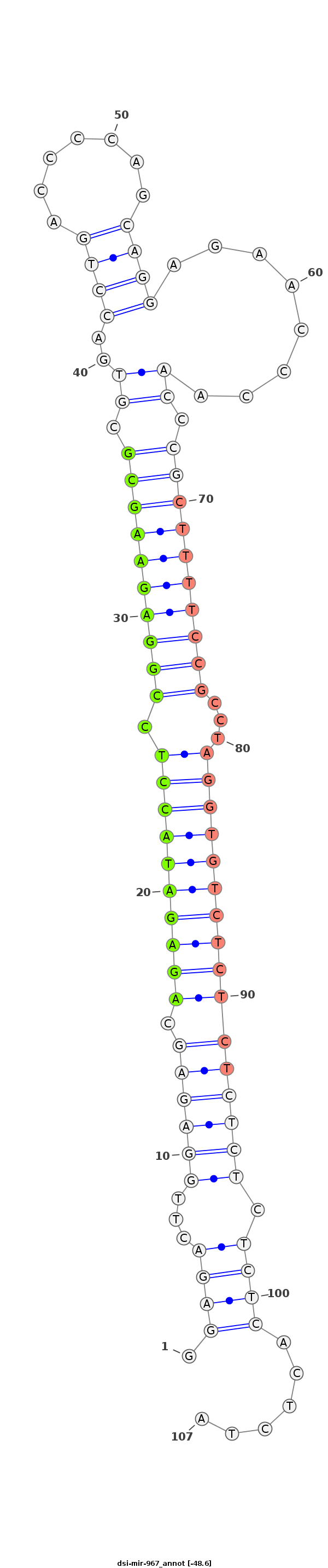
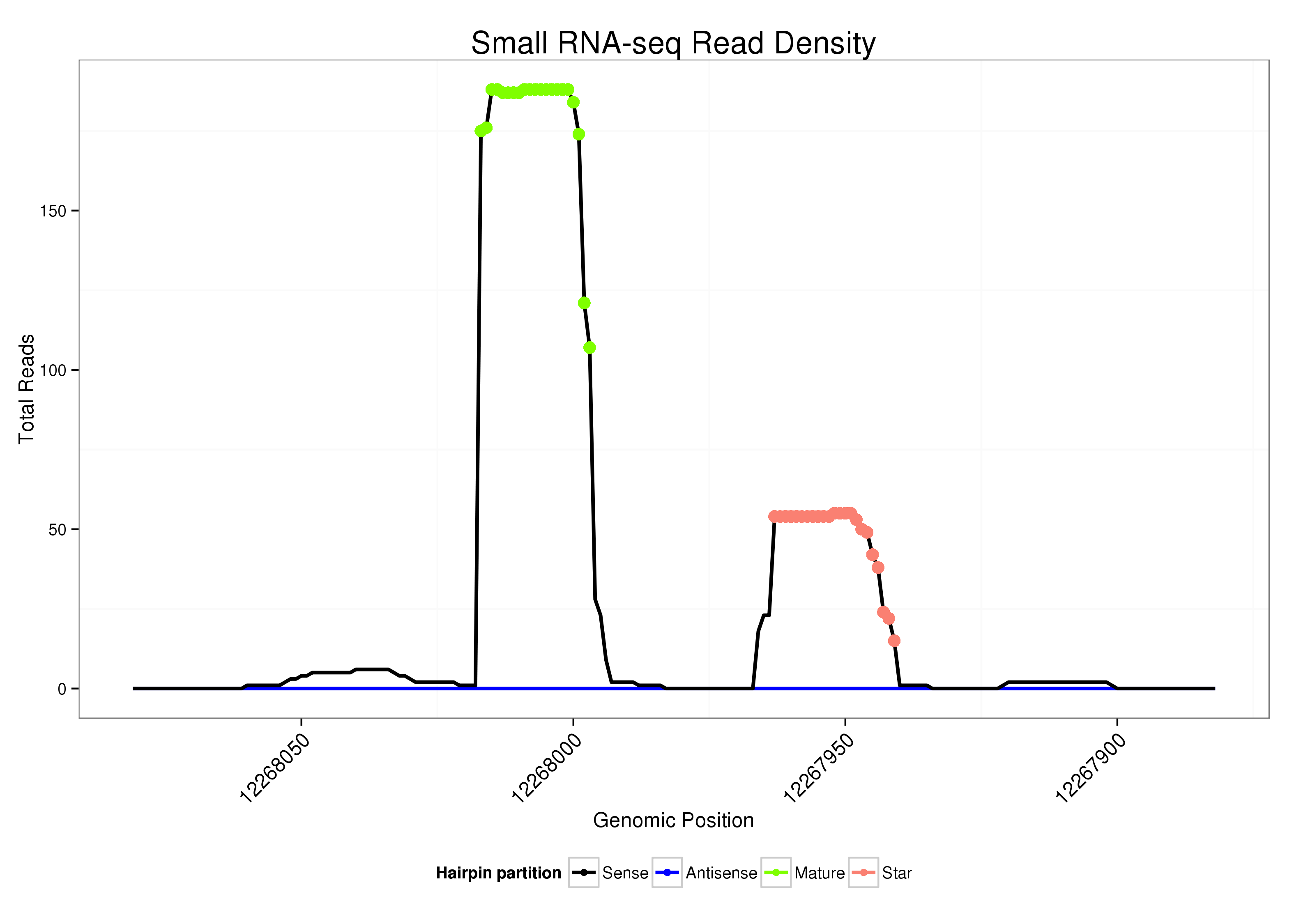
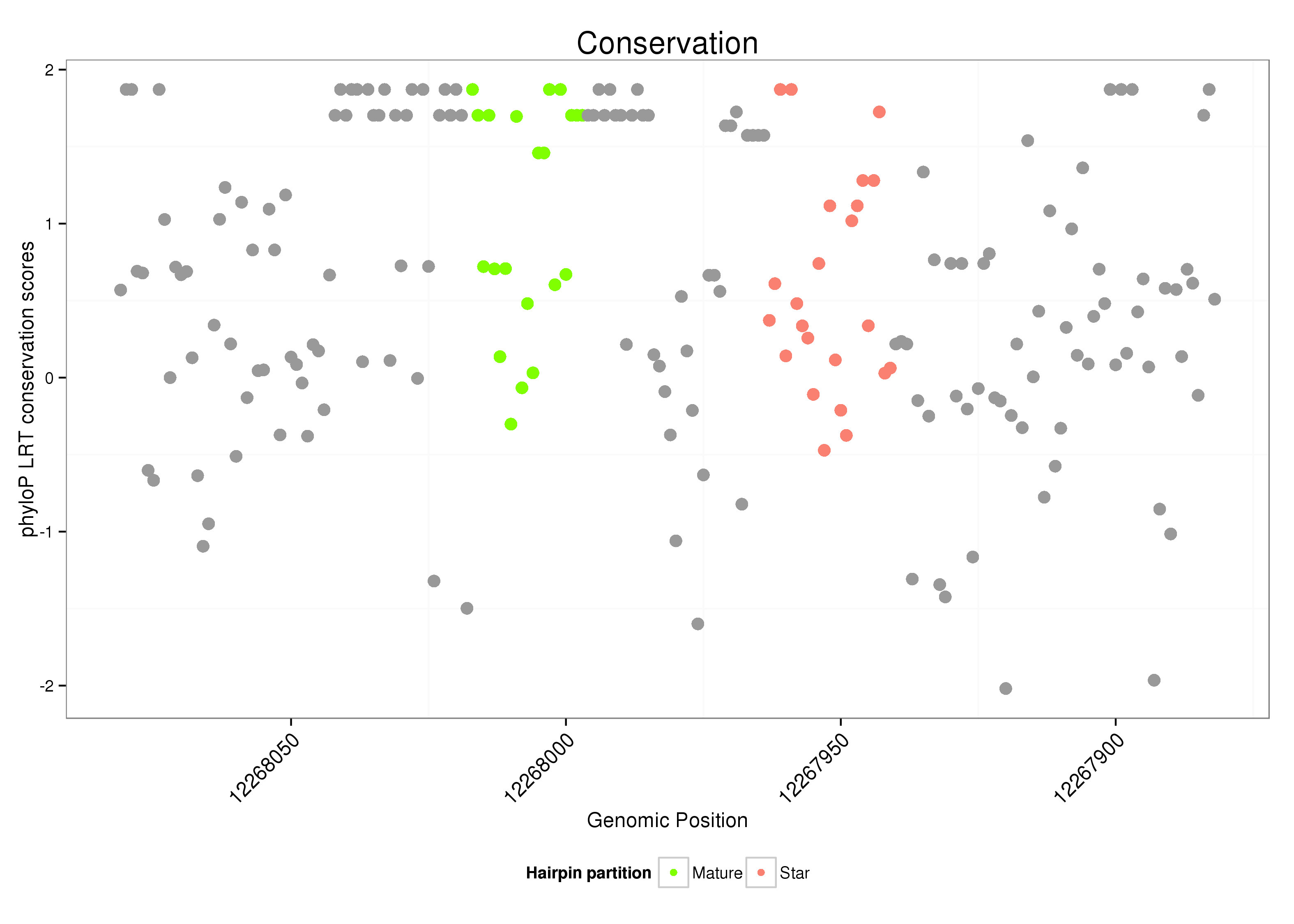
ID:dsi-mir-967 |
Coordinate:2L:12267932-12268031 - |
Confidence:Known |
Type:Unknown |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
intron [Dsim\GD22116-in]
No Repeatable elements found
|
GAAAACCAAAAAAAAAAAAACACCAAGAAAACCAAAACTGAGAACTGCAGGAGACTTGGAGAGCAGAGATACCTCCGGAGAAGCGCGTGACCTGACCCCAGCAGGAGAACCCAACCCGCTTTTCCGCCTAGGTGTCTCTCTCTCTCTCTCACTCTATCTCAGTTCCTCAGCTCGAGAGTGTGTATCTTACCCCGAGTGTG
*************************************************.((((...((((((.((((((((((.((((((((((.((..((((.......))))........)).))))))))))...)))))))))))))))).))))......******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O002 Head |
M024 male body |
GSM343915 embryo |
M025 embryo |
M053 Female-body |
SRR618934 Ovary |
O001 Testis |
SRR553485 Ovary |
SRR553486 Ovary |
SRR553487 Ovary |
SRR553488 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................AGAGATACCTCCGGAGAAGCG................................................................................................................... | 21 | 0 | 1 | 78.00 | 78 | 38 | 0 | 2 | 10 | 8 | 6 | 5 | 0 | 4 | 1 | 1 | 3 |
| ................................................................AGAGATACCTCCGGAGAAG..................................................................................................................... | 19 | 0 | 1 | 53.00 | 53 | 29 | 1 | 22 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTCC........................................................... | 23 | 1 | 1 | 17.00 | 17 | 10 | 0 | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTCT........................................................... | 23 | 0 | 1 | 14.00 | 14 | 8 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGCGCG................................................................................................................. | 23 | 0 | 1 | 13.00 | 13 | 0 | 9 | 0 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGC.................................................................................................................... | 20 | 0 | 1 | 12.00 | 12 | 4 | 4 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTC.............................................................. | 20 | 0 | 1 | 10.00 | 10 | 6 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAA...................................................................................................................... | 18 | 0 | 1 | 10.00 | 10 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGCGCGT................................................................................................................ | 22 | 0 | 1 | 7.00 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 2 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGTC................................................................ | 21 | 0 | 1 | 6.00 | 6 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTC............................................................ | 22 | 0 | 1 | 6.00 | 6 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGCGT.................................................................................................................. | 22 | 1 | 1 | 6.00 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGTCT............................................................... | 22 | 0 | 1 | 4.00 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGCGC.................................................................................................................. | 22 | 0 | 1 | 4.00 | 4 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGA....................................................................................................................... | 17 | 0 | 1 | 4.00 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| ....................................................................................................................CGCTTTTCCGCCTAGGTG.................................................................. | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGT................................................................... | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CGCTTTTCCGCCTAGGTGTCTC.............................................................. | 22 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGTCTC.............................................................. | 23 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCTGGAGAAGCG................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAGGCG................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGCGA.................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGTCTCTC............................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTTTCTCTC............................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGT................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGCGCGTGACCT........................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGCGCG................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAGG.................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGCG................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................ACCAAGAAAACCAAAACTGAGAACTGC........................................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTCTCTCTCTA.......................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................TCCGGAGAAGCGCGTGACCTGAA........................................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTGTCTCT............................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................CCAAAACTGAGAACTGCAGG..................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AGGTGTCTCTCTCTCTCT..................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CGCTTTTCCGCCTAGGTGTC................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................AGTTCCTCAGCTCGAGAGTGT................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................GAGATACCTCCGGAGAAGC.................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTTTCCGCCTAGGTGTCTCTCC........................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCT............................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................CCGCTTTTCCGCCTAGGTG.................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................GAACTGCAGGAGACTTGGAGAGCAGAG.................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGTTGTCT............................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................AAAACTGAGAACTGCAGGAGACTTGGA............................................................................................................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGCGC.................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGTTGTCTCTC............................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGCAGCGC.................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................AAACCAAAACTGAGAACTGCA....................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................CTCCGGAGAAGCGCGTGACCTGACCC...................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTCCT.......................................................... | 24 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CCGGAGAAGCGCGTGACCTGG......................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGATAAGC.................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................AACCAAAACTGAGAACTGCAGGA.................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTCG........................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................GTTCCTCAGCTCGAGAGTG.................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AGAGATACCTCCGGAGAAC..................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGATACCTCCGGAGAAGC.................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTTTTCCGCCTAGGTGTCTCTCTT.......................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................TCCTGAGGTCGAGAGT..................... | 16 | 2 | 20 | 0.50 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 6 |
| .............................................TGCAGGACACTTGGGGAGCA....................................................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................GAGCGCGGGACCTGAACC...................................................................................................... | 18 | 3 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 0 |
| ...............................................................................GGAGCGCGGGACCTGAACC...................................................................................................... | 19 | 3 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| .................................................................................AGCGCGGGACCTGAACC...................................................................................................... | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................CGAACCCGTTTTCCCGCCTA...................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................GGAGGACCGAACCCGC................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................AGCGCGGGACCTGAACCG..................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................AAGTAGCTCCGGAGAAGC.................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CACACTCGGGGTAAGATACACACTCTCGAGCTGAGGAACTGAGATAGAGTGAGAGAGAGAGAGAGACACCTAGGCGGAAAAGCGGGTTGGGTTCTCCTGCTGGGGTCAGGTCACGCGCTTCTCCGGAGGTATCTCTGCTCTCCAAGTCTCCTGCAGTTCTCAGTTTTGGTTTTCTTGGTGTTTTTTTTTTTTTGGTTTTC
********************************************......((((.((((((((((((((((...((((((((((.((........((((.......))))..)).)))))))))).)))))))))).))))))...)))).************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 Ovary |
SRR553487 Ovary |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|
| .......................................................................AGGCGAAAAAGCGTGTTG............................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| ......................................................................................................................................................................TGGTTGTCTTGGAGTTTTTTT............. | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ............................................TAGAGTGAGAGAGAGAG........................................................................................................................................... | 17 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...................................................................................................................................................................TTTTGGTTTTCTTGGT..................... | 16 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| .........................................................................................................................................................................TTTTCTTCGTGTTTTTTTTTGT......... | 22 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 |
| .............................................AGAGTGAGAGAGAGAGAGA........................................................................................................................................ | 19 | 0 | 16 | 0.06 | 1 | 0 | 0 | 1 |
| ..................................................GAGAGAGAGAGAGAG....................................................................................................................................... | 15 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 2L:12267882-12268081 - | dsi-mir-967 | GAAAACC-------------AAAAAAA-A-AAAA-----ACACCAAGAAAACCAAAAC-----TGAGAACTGCAG--GAGA------CT-TGGAG-----------------------AGCAGAG---------ATA-------------------------------------CCTCCG----------------GAGAAGCGCGTGACCTGACCCCAGCAGG----------------AG-AACCCAACCCGCTTTTCC--------G-CCT-----------------AGGTGTCTC-TCTCTCTCTCT--C--------AC--TCTATCTCAGTTCCT------CAGCTCGAG----------AGTGTGTATCTTA-CCCCGAG--TGTG |
| droSec2 | scaffold_16:650994-651190 - | dse_56 | GAAAACC-------------AAAAAAA-AAAAAA-----ACACCAAGAAAACCAAAAC-----TGAGAACTGCAG--GAGA------CT-TGGAG-----------------------AGCAGAG---------ATA-------------------------------------CCTCCG----------------GAGAAGCGCGTGACCTGACCCCAGCAGG----------------AG-AACCCAACCCGCTTTTCC--------G-CCT-----------------AGGTGTCTC-TCT----CT--CTC--------TC--TCTATCTCAGTTCCT------CAGCTCGAG----------AGTGTGTATCTTA-CCCCGAG--TGTG |
| dm3 | chr2L:12459963-12460153 - | dme-mir-967 | AAAAACC------------GAA-AAAA---AAAA-----ACACTAAGAAAACCAAAAA-----TGAGAACTGCAG--GAGA------CT-TGGAG-----------------------AGCAGAG---------ATA-------------------------------------CCTCTG----------------GAGAAGCGCGTGACCTGACCCCAGCAGG----------------AG-AACCCAACCCGCTTTTCC--------A-CCT-----------------AGGTGTCTC-TCT----CT----C--------TC--TTTATCT--GTTCCT------CAGCTCGAG----------AGTGTGTATCTTT-CCCCGAG--TGTG |
| droEre2 | scaffold_4929:12958907-12959115 + | GAAAAGCGA-----AAACCGAA-AAAA---CGAA-----AAACCAAGAAAACCAAAAC-----TGAGAACTGCAG--GAGA------CT-TGGAG-----------------------AGCAGAG------ATGATA-------------------------------------CCTCTG----------------GAGAAGCGCGTGACCTGACCCCAGCAGG----------------AG-AACCCAGCCCGCTTTTCC--------G-CCT-----------------AGGTGTCTC-TCACTCTCTCT--C--------TC--TCTCTCTCTTCTCCT------CAGCCCGAG----------TGTGTGTATCTTT-CCCCGAA--TGTG | |
| droYak3 | 2L:8897045-8897266 - | GAAAACCGAAACCAAAACCGAA-A--------AA-----AAACCAAGAAAACCAAAAC-----TGAGAACTGCAG--GAGACGGAGACT-TGGAG-----------------------AGCAGAGACAGAGATGATA-------------------------------------CCTCTG----------------GAGAAGCGCGTGACCTGACCCCAGCA-------------------G-AACCCAACCCGCTTTTCC--------G-CCT-----------------AGATGTCTC-TATCTCGCTCT--C--------CC--TCTCTCT--GTTCCTCTTGCACAGCTCGAG----------TGTGTGTATCTTT-CCCCGAG--TGTG | |
| droEug1 | scf7180000409122:844094-844271 - | GAACAAA-------------A----AACAC---A---------------CATCGAAAC-----TGAGAACTGCAGAGGAGA------CT-TTGAGAAG--------------------AGAAGAG---------ATA-------------------------------------ACACTG----------------GAGAAGCGCGTGACCTGACCCCAGAAGG-----------------C-AACCCAACCCGCTTCTCC-----AG-G-CCA-----------------AGAGGTCTCCTCT----CT--CTC--------GC--TCGCTCTCACTCCC--------------------------TGTGTTTTTCTCA-GACCAAG--TGTG | |
| droBia1 | scf7180000302422:507050-507210 + | GAAAATC-------------A----G--C-------------------------AGGC-----TGAGAACTGCAG--GAGA------CT-CCGAG-----------------------AGGAGAG-CGGAGA-GATA-------------------------------------CCTCTG----------------GAGAAGCGCGTGACCCGACCCCGAG--G----------------GC-TGCCCAGCCCGCTTCTCC-----AGAGGG-C-----------------AGAGGTCGC-TCT----CT--GTG------------------------CCT------CTGCC---G----------CGTGTGTAT--CC-CCACGAG--CGTG | |
| droTak1 | scf7180000415865:119221-119370 + | GAAAAAA-------------A----AC-C-------------------------GAAC-----TGAGAACTGCAG--GAGA------CT-TGGAG-----------------------AGAAGAG---------CTA-------------------------------------TCTCTG----------------GAGAAGCGCGTGACCTGACCTGAG----------------------------AACCCGCTTCTCC-----AG-G-G-C-----------------AGAGGTCTC-TCT----CT--AT------------------------TTCT------CAGACCGAG----------TGTGTGTATCTCCCCCTCGAG--TGTG | |
| droEle1 | scf7180000491028:2648335-2648512 + | GAAAAAA-------------A----AA-C-------------------------GAAC-----TGAGAACTGCAT--GGGACTGAGACT-TCGAG-----------------------AGCAGAG---------ATA-------------------------------------CCTCTG----------------GAGAAGCGCGTGACCCGACCTCAGAAGG-----CACCAGCCAACCC-AATCCAACCCGCATCTCC------------------------------AGACGTCTC-TCT----T-----------------------------TTCT------CCGCCCGAGTGTGTGTG-GTGTGTGTATCTCG-TCCCGAG--TGTG | |
| droFic1 | scf7180000454015:349245-349421 - | GAAAAAA-------------A----AT-CA---AAATCAAAGC------------GAC-----TGAGAACTGCAG--GAGA------CTTTCGAG-----------------------AGCAGGG---------ATAT------------------A-----------------TCTCTG----------------GAGAAGCGCGTGACCTGACCCCAGAAGT----------------AGCAACCCAGCCCGCTTCTCC-----GG-G-CCA-----------------AAAGGTCTC-TCT----TT----T--------CCTTGCGGCCTTAAT------------------G----------TGTGTGTATCTCC-CCCCGAG--AGTG | |
| droKik1 | scf7180000302472:819802-820012 - | GAAACCG-------------AA-AAAATAA---A---------------AACC-GAAA-----TGAGAACTGCATATGAGA-TGA--TTTTCGAGAAGCGAGAAACGAGAAAACCGAGAGGAGAG---------ACGT------------------GTTTTTTGTTGTGAGATACCTCAGTTTAGGAGAAGGCAAAGAAAAGCGCGTGACCTGACCCCATGA-------------------A-AACCC--------TCTCC-----AC----------------------------------------------A------------------------ACA------C--GCT--GTGTGTGTG-CTGTGTGTATCTCG-GCGAGAGTGTGTG | |
| droAna3 | scaffold_12916:12931155-12931336 + | AAAAAAA-------------A----AA-A-------------------------AAACTCATCCGAGAAATGCAT--GAGA------TT-TTGAG-----------------------AGGAGAG---------ATA-------------------------------------CCTC-------------------AGAAGCGCGTGACCTGACCCCGTCCGTCTGTCCATCTA-----TG-AACCCAACCCGCTTTTCT---CCAAACAC-CAGAGAGGAGGTCGAGAG----------TG--------------------------------------A------C------GGTGTGTGTGAGTGTGTCTCTCTCCCCCTCAAG--TGTC | |
| droBip1 | scf7180000396580:589804-589961 - | GAAAAAT-------------A----AA---------TCCAACCG-AGA-------ACA-----TGAGAAGTGCAT--GAGA------TT-TTGAG-----------------------AGGAGAG---------ACA-------------------------------------CCTC-------------------AGAGGCGCGTGACCTGACCCGT-CA-G----------------CG-AACCCAGCCCGCTTTTCTC-----------------------------------CTC-TCC----T----CT------------------------CCT------C--TCCGGGTGTGTGTG--TATGTGTCTCTCC-CCTCAAG--AGTC | |
| dp5 | 4_group3:3150540-3150765 - | GAAAAAA-------------A----AA-CAT-----TCAAAACGAGAAAGACAAAAAT-----TGAGAACTGCAT--GAGA------CT-TGGAG-----------------------AGAAGAG---------ATACCCACAAGAGGCAGGCAGTG------------A------AGTG---GAGTGGAGGGAGCGAGAAGCGCGTGACCTGACCCCAAGAC-----------------CG-AACCCAACCCGGTTCTGTTCTCCAGTGTA-CAGACAGGCA-----------------CTCT----GTAT--GAGTATGTAC------------CCTCCT------TCGCTC---------------CCTGTATGCGT-GCACGTG--TGTG | |
| droPer2 | scaffold_1:4633223-4633444 - | GAAAAAA-------------AA---AA-CAT-----TCAAAACGAGAAAGACAAAAAT-----TGAGAACTGCAT--GAGA------TT-TGGAG-----------------------AGAAGAG---------ATACCCACAAGAGACGGT-----------------------CCGTG---GAGTGGAGGGAGCGAGAAGCGCGTGACCTGACCCCAAGAC-----------------CG-AACCCAACCCGGTTCTGTTCTCCAGTGTA-CAGACAGGCA-----------------CTCT----GTAT--GAGTATGTAC------------CCTCCT------TCGCTC---------------CCTGTATGCGT-GCACGTG--TGTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 12/06/2013 at 02:36 PM