| droSim3 |
3L:12997814-12998009 + |
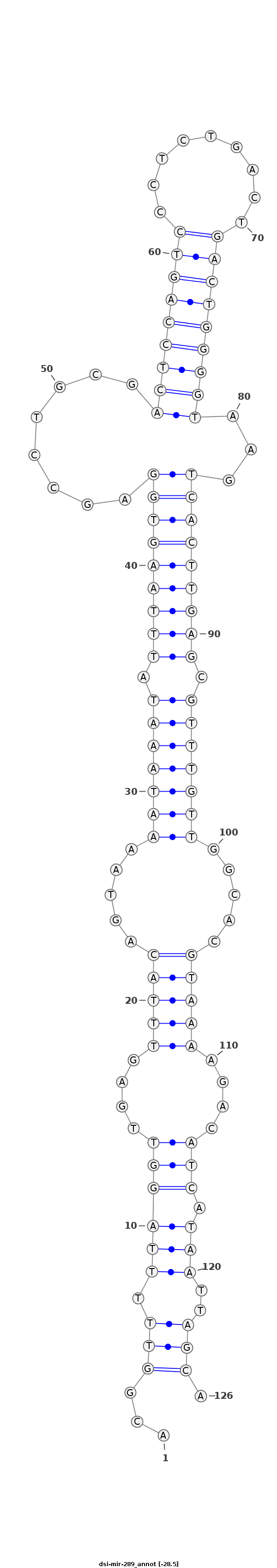
dsi-mir-289 |
GGCA------------GTCGT-------------CGTAGTCGCAGTC----------------------------------------GCAGTCGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------C-------------------------------------------------------------------------------------AGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTTGAAATTAGTGCGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| droSec2 |
scaffold_0:5775552-5775747 + |
dse-mir-289 |
GGCA------------GTCGT-------------CGTAGTCGCAGTC----------------------------------------GCAGTCGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------C-------------------------------------------------------------------------------------AGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTTGAAATTAGTGCGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| dm3 |
chr3L:13613867-13614068 + |
dme-mir-289 |
GGCA------------GTCGT-------------CGTAGTCGCAGTCGTAG------TC----------------------------GCAGTCGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTT---------C-------------------------------------------------------------------------------------AGTCCCTCTGACTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTTGAAATTAGTGCGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| droEre2 |
scaffold_4784:13614815-13615016 + |
der-mir-289 |
GGCA------------GTCGT-------------CGCTGTCGCAGTCGCAG------TC----------------------------GCAGTCGTC-----------------------------------------------------------------------GTCAACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------C-------------------------------------------------------------------------------------AGTCCCTCTGACGGACTGGGGTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGCGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| droYak3 |
3L:13706908-13707115 + |
dya-mir-289 |
GGCA------------GTCGT-------------CGTAGTCGCAGTCGCAGTCGGAGTC----------------------------GCAGTCGTC-----------------------------------------------------------------------GTCTACGGTTCTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------C-------------------------------------------------------------------------------------AGTCCCTCTGACTGACTGGGGTAAGTCTCTCGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGCGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| droEug1 |
scf7180000409121:150404-150590 + |
|
GGCA------------GCAG--------------------------------------------------------------------CAGCAGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTCG-------------------------------------------------------------------------------------CTTTCTCTCTCTCTCTCTCTTTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGCCAGC---------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000302428:6302573-6302745 - |
|
GCCA------------GTCGT-------------AGTC-------------------------------------------------ATGGTAGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGTGACCGCG---ACGAC-------------------------------------------------------------------------------------TC-----CCCT-CCACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGT--------------------------------------------------------------------------------------------------- |
| droTak1 |
scf7180000415161:62302-62461 + |
|
GGCG---------GCAGTCG----------------------------------------------------------------------------------------------------------------------------------------------------TCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------C---------------------------------------------------------------------------------------------TTG-CGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGCC------------------------------------------------------------------------------------------- |
| droEle1 |
scf7180000491255:2056090-2056264 + |
|
GGCA------------GTCGA-------------CGTCGT----------------------------------------------------CGTC-----------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTGAGTGGAGCCTGCGACTC---------C---------------------------------------------------------------------------------------------TTGCCGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGCCAGC-------------------------------------------------------------------------------------GA- |
| droRho1 |
scf7180000779509:202430-202598 + |
|
GGCA------------GTCG----------------------------------------------------------------------------------------------------------------------------------------------------TCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTGAGTGGAGCCTGCGACTC---------C---------------------------------------------------------------------------------------------TTGCCGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGCCAGCGAGTG--------------------------------------------------------------------------------AGC |
| droFic1 |
scf7180000454048:90324-90523 - |
|
GGCA------------GTCGT-------------C------------------------------------------------------------------------------------------------------------------------------------GTCTACGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------CTTCCCTTCCCTTCCCTTCCCTTCCCTTCCCTTCCCT---------------------------------------------------------TCGCTA---AGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGCCAGC-------------------------------------------------------------------------------------AGC |
| droKik1 |
scf7180000302808:404312-404492 - |
|
GGCA------------GTCGT-------------CGGCGTT-----CCTTG------TC----------------------------CCTGCCGCC-----------------------------------------------------------------------GTCTAGGGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC-------------------------------------------------------------------------------------------------------TGGCTGACTGGGGTAAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGCCATGC-------------------------------------------------------------------------------------------- |
| droAna3 |
scaffold_13337:21827545-21827780 - |
dan-mir-289 |
GGCA------------GTCGT-------------TGGCGTT-----TCT----------GGTCCTTTCTCCTTTCTCCTTCTTCCGG---------CACTCCGGCA--------------------------------------------------------TTCC-AGC-TCGGGTTTTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC---------TG-----------------------------------CTACTGCC---ACTGCCACTGCCACTGCCAC---------------------------TGCCGCT--CGGGGAGTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGC-------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396730:303826-304034 - |
|
GGCA------------GTCGT-------------TGGCGTT-----TCT----------GGT------------CTCCTTCT-CCGG---------CACTCCGGCACT----------------------------------------CCAGTACTCTGGCATTCC-GGC-TCGGGTTCTAGGTTGA-GTTTACAGTAAAATAAATATTTAAGTGGAGCCTGCGACTC--------------------------------------------------------------------------------------------------------TG-CCACTGGGG-GAGTCACTTGAGCGTTTGCTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGTGTGGCATGC-------------------------------------------------------------------------------------------- |
| dp5 |
XR_group8:6818863-6819118 - |
dps-mir-289 |
GGCA------------GTCGT-------------CCTCGTCCTCGTCCTCG------TC----------------------------CTAG-----CACCGCTCCGCTCCGTACCACACCGTACCGTTCAGTTCAGTTCAGTTGGGAACCAGTAC----GTATATCC--GTCGGGTTTTTAGGTTGA-GTTTACAGTGAAATAAATATTTAAGTGGAGCCTGCGACTGGG---ACTCC-------------------------------------------------------------------------------------AG-----CTCTCCGACTGGGCTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGCTTGACATGC-------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_36:488052-488301 + |
dpe-mir-289 |
GGCA------------GTCGT-------------CCTCGTCCTCGTC----------------------------------------GTAG-----CATCGCTCCACTCCGTACCACACCGTACCGTTCAGTTCAGTTCAGTTGGGAACCAGTAC----GTATATCC--GTCGGGTTTTTAGGTTGA-GTTTACAGTGAAATAAATATTTAAGTGGAGCCTGCGACTGGG---ACTCC-------------------------------------------------------------------------------------AG-----CTCTCCGACTGGGCTAACTCACTTGAGCGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGC--GCTCGAAATTAGCTTGACATGC-------------------------------------------------------------------------------------------- |
| droWil2 |
scf2_1100000004511:607044-607331 - |
|
--CA------------GTCGT-------------CGACG-------------------------------------------------------GT-----------------------------------------------------------------------TTCTTTTTTTTTTAGGTTGAAGTTTACAGTAAAATAAATATTTAAGTGGAGTC---GACTGTGTCGACTAC----------------------------------------AGCCCGGACTACC---CGGACTGCTAGAACGACTCGGTTC-----CGACTCTTTTTCTGTCGGGGTAAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCATTGGGCGCGCTGAAAATTAGTGTAGCATACT-----------------------TACTGGCTGCCTGACTGCCGAGATGCCAGAGCAGCAGTAGCAGCAGCAGTAGCAGCAGCAGTAGCAGC |
| droVir3 |
scaffold_13049:19817922-19818108 - |
|
TGCG-TTTCTGCGG-------------CAGTCGACGTCGT-------------------------------------------------------------------------------------------------------------------------------------GTTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGG-GCCT------------------------------------------------------------------------------------------------------------------------GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGTTCG-----TCGAAATTAGTGCGGCGGCCCAAAAAAAGAAAACGTAGCATACTTTTTGTCG--------------------------------------------------CA-TAG--CC |
| droMoj3 |
scaffold_6680:7513449-7513598 - |
|
AACG-TT--TGC---AGTCGTCGCTGGCAGTCGACG---------------------------------------------------------------------------------------------------------------------------------------TGGCTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGGGCCC-------------------------------------------------------------------------------------------------------------------------GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGTTGGC-----TGGAATTAGAGCGGC------------------------------------------------------------------------------------------------ |
| droGri2 |
scaffold_15110:17555322-17555471 + |
|
TGTCGTT--TGCGG-------------CAGTCGACG---------------------------------------------------------------------------------------------------------------------------------------TCAGTTTTTAGGTTGA-GTTTACAGTAAAATAAATATTGAAGTGG-GCCT------------------------------------------------------------------------------------------------------------------------GGGGAAGTCACTTGAACGTTTGTTGGCACGTAAAAGACATCATAATTAGCCGTTCG-----TTCAAATTAGTGCGGCGGCCCAAC-------------------------------------------------------------------------------------A-- |