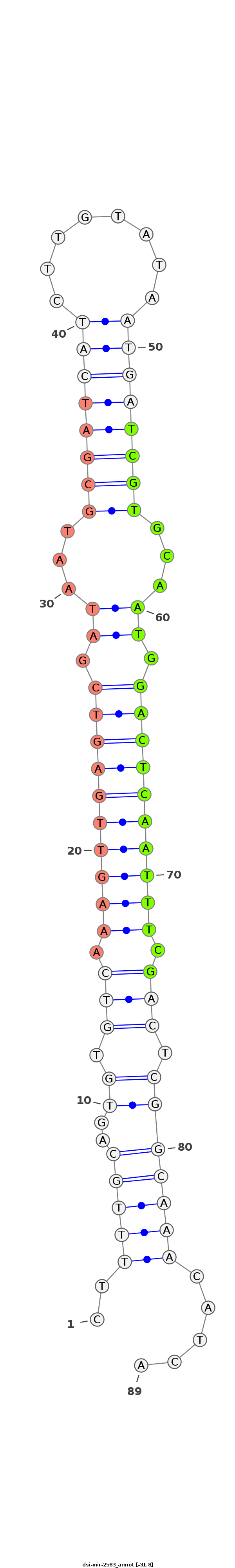
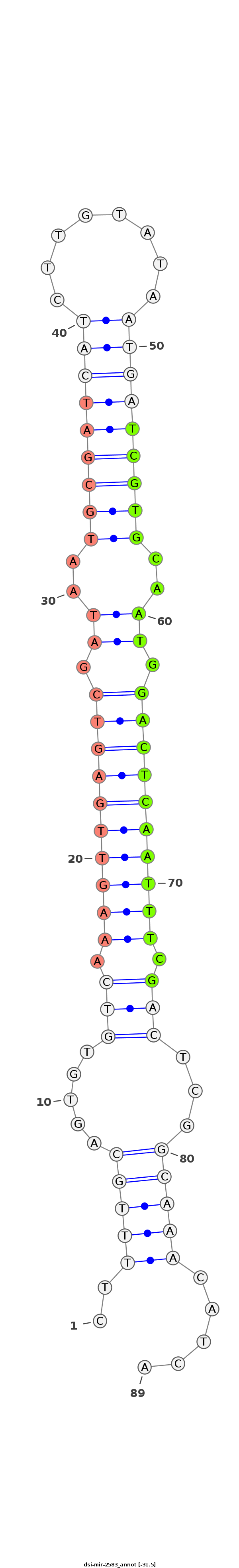
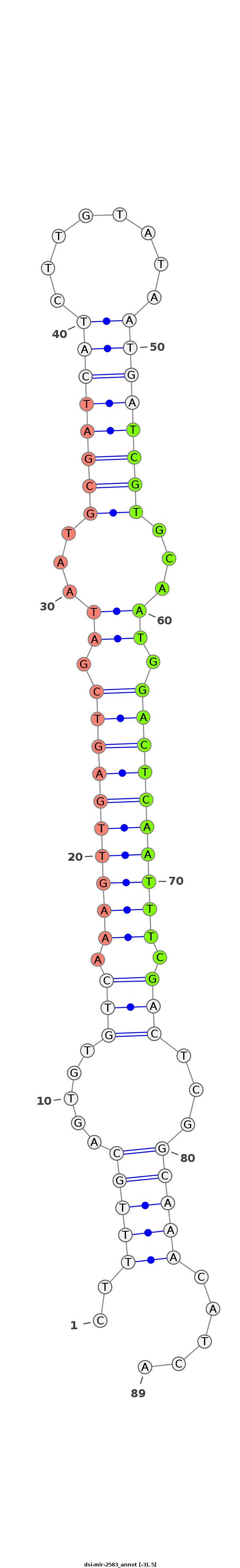
| ....................................................................................................TCGTGCAATGGACTCAATTTCG............................................................ |
22 |
0 |
1 |
73.00 |
73 |
35 |
10 |
6 |
9 |
0 |
8 |
4 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGAT................................................................................................. |
22 |
0 |
1 |
37.00 |
37 |
7 |
1 |
5 |
4 |
11 |
0 |
4 |
0 |
2 |
0 |
3 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGA.................................................................................................. |
21 |
0 |
1 |
26.00 |
26 |
6 |
3 |
7 |
0 |
0 |
4 |
2 |
2 |
1 |
0 |
1 |
0 |
| ....................................................................................................TCGTGCAATGGACTCAATTTC............................................................. |
21 |
0 |
1 |
12.00 |
12 |
7 |
4 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGATC................................................................................................ |
23 |
0 |
1 |
4.00 |
4 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGC.................................................................................................... |
19 |
0 |
1 |
4.00 |
4 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
1 |
0 |
0 |
| .....................................................................................................CGTGCAATGGACTCAATTTCGA........................................................... |
22 |
0 |
1 |
3.00 |
3 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................................GTTGAGTCGATAATGCGATC................................................................................................ |
20 |
0 |
1 |
3.00 |
3 |
2 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCG................................................................................................... |
20 |
0 |
1 |
3.00 |
3 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................CGTGCAATGGACTCAATTTCG............................................................ |
21 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................TCGTGCAATGGACTCAATTTCGG........................................................... |
23 |
1 |
1 |
2.00 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................TCGTGCAATGGACTCAAT................................................................ |
18 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGATA................................................................................................ |
23 |
1 |
1 |
2.00 |
2 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................AAGTTGAGTCGATAATGCGATC................................................................................................ |
22 |
0 |
1 |
2.00 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ....................................................................................................TCGTGCCATGGACTCAATTTC............................................................. |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ..................................................................GTTGAGTCGATAATGCGGTGCT.............................................................................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ....................................................................................................TCGTGCAATGGACTCAATTTCGA........................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................CAGTTGAGTCGATAATGCGATC................................................................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................GATCGTGCAATGGACTCAAT................................................................ |
20 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................................TCGTGCAATGGACTCAACTTCG............................................................ |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................TGCAGTGTGTCAAAGTTGAGT............................................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGAC................................................................................................. |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................CGTGCAATGGACTCAATTTC............................................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGATT................................................................................................ |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ....................................................................................................CCGTGCAATGGACTCAATTTCG............................................................ |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................TGAGATAATCTTGTATGATGAT................................................................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................................GTTGAGTCGATAATGCGATCA............................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGTAA................................................................................................ |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATGCGG.................................................................................................. |
21 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................AAAGTTGAGTCGATAATCCGAT................................................................................................. |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................ATCGTGCAATGGACTCAATTTCG............................................................ |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................GTTGAGTCGATAATGCGATTAT.............................................................................................. |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................................................................ATCGTGCAATGGACTCAATTT.............................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................TTGAGTCGATAATGCGAT................................................................................................. |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................CGTGCAATGGACTCAATTTCGT........................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................GTTGAGTCGATAATGCGA.................................................................................................. |
18 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................ATCGTGCAATGGACTCAATTTC............................................................. |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................TGAGATCATTTTGTATGATGAT................................................................................. |
22 |
3 |
6 |
0.67 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
4 |
0 |
0 |
0 |
| ...............................................................................TGTGATCATTTTGTATATTGAT................................................................................. |
22 |
3 |
5 |
0.20 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...............................................................................TGCGATCATTTTGTATGCTGAT................................................................................. |
22 |
3 |
5 |
0.20 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................................TGACCGTGGAATGAACTCA.................................................................. |
19 |
3 |
16 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................GTGGCCGTCTCTTTT................................................................................................................................. |
15 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |