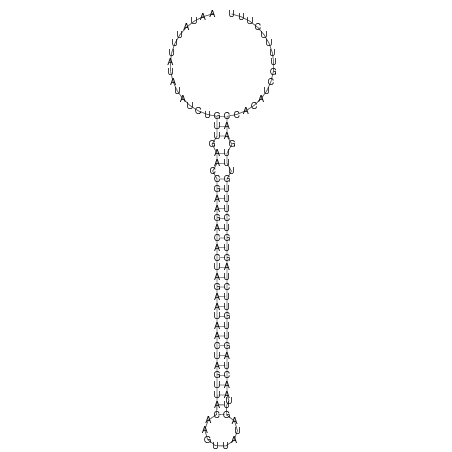
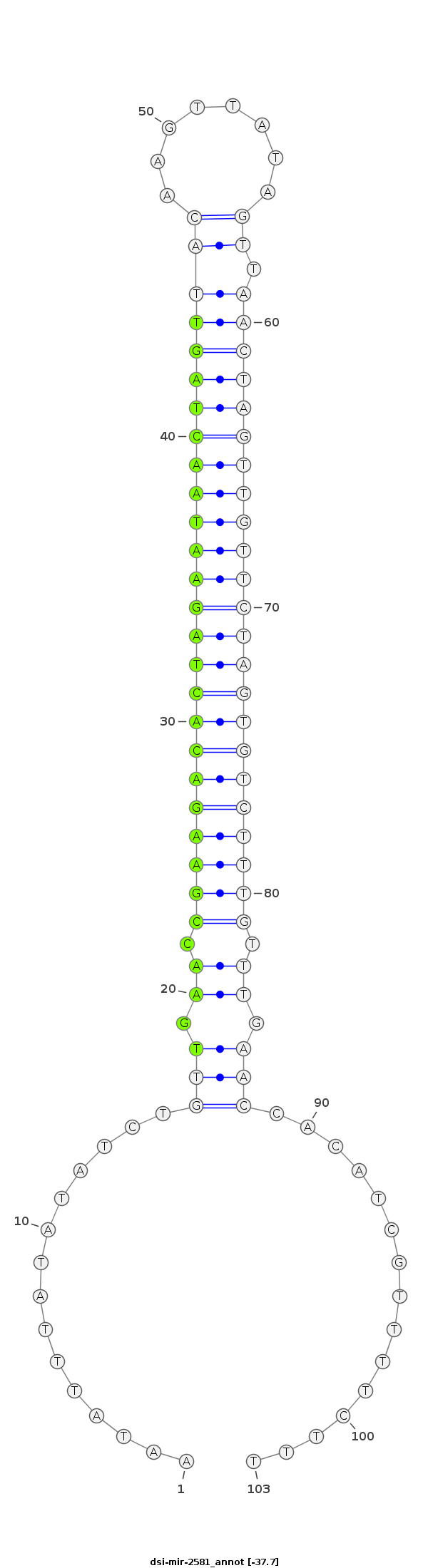
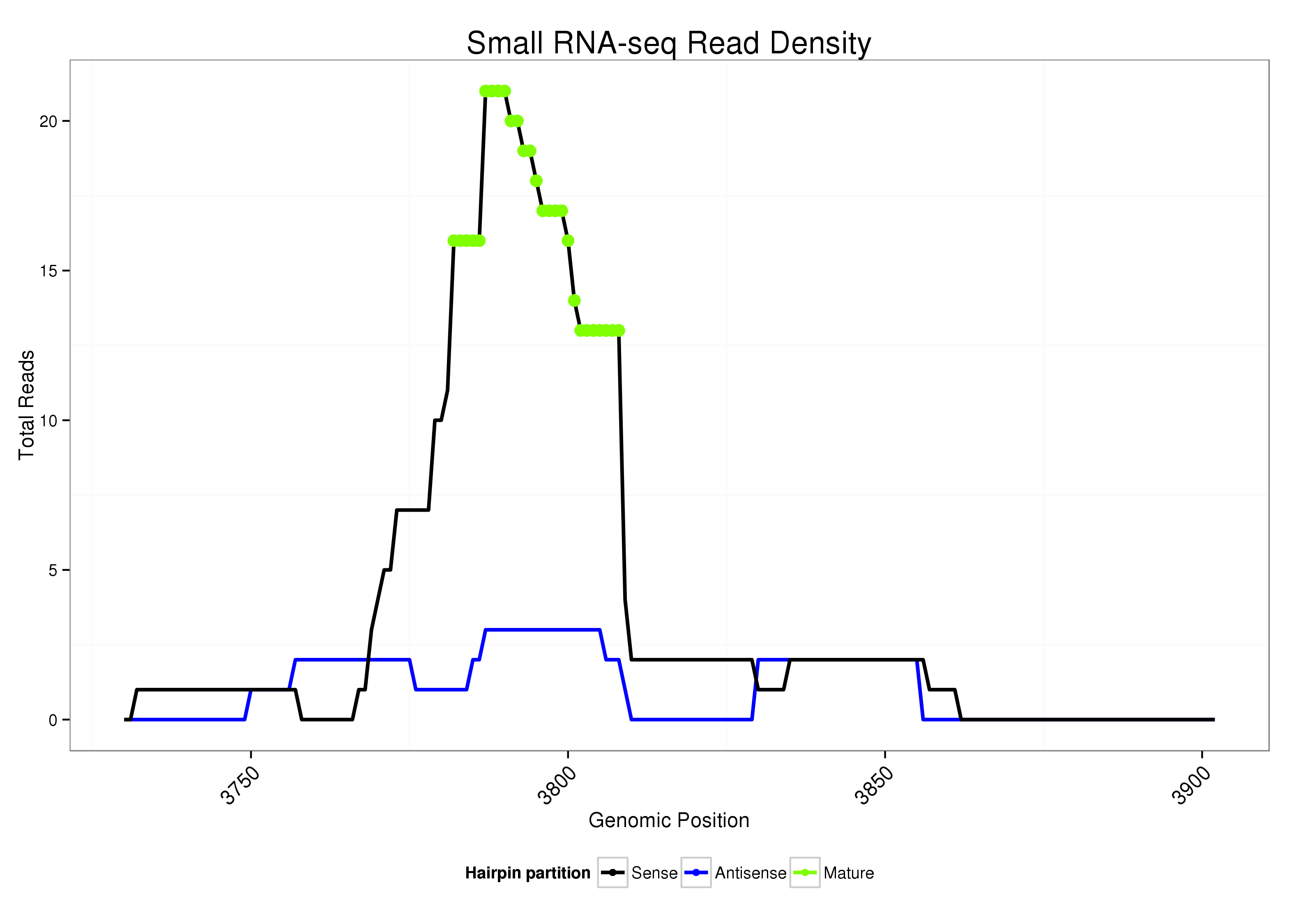

ID:dsi-mir-2581 |
Coordinate:chrXh_Mrandom_005:3780-3852 + |
Confidence:Known |
Type:Unknown |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -37.0 | -36.9 |
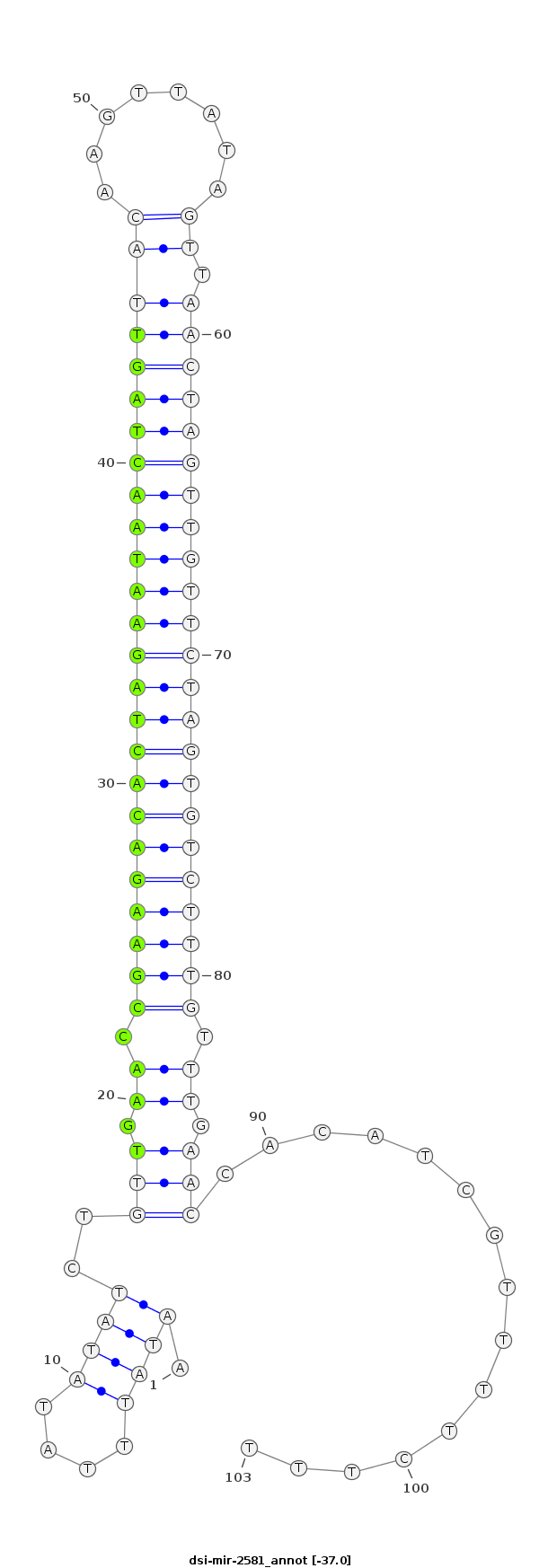 |
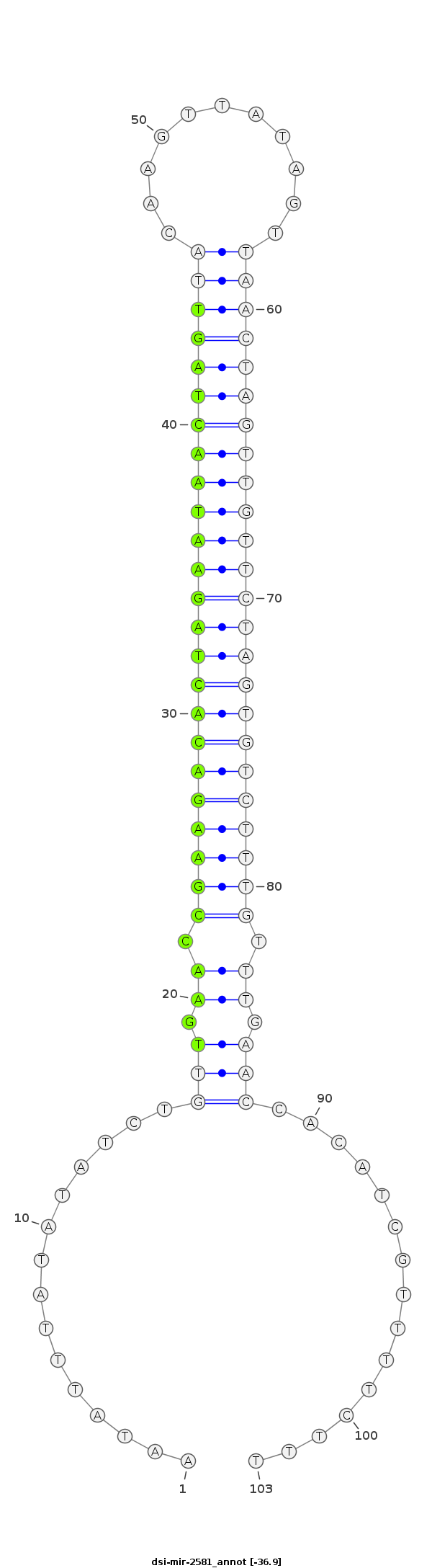 |
intergenic
No Repeatable elements found
|
ATTTGTCTGCACTAATAAGTTTTTCCCTTGATACAAATATTTATATATCTGTTGAACCGAAGACACTAGAATAACTAGTTACAAGTTATAGTTAACTAGTTGTTCTAGTGTCTTTGTTTGAACCACATCGTTTTCTTTTTTTTTCATTCTGCAAAATGAGTTTTATTCAGTGT
***********************************...............(((.((.(((((((((((((((((((((((((........)).))))))))))))))))))))))).)).)))...............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Ovary |
GSM343915 embryo |
SRR553487 Ovary |
SRR618934 Ovary |
SRR553488 Ovary |
SRR553486 Ovary |
M025 embryo |
M053 Female-body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................TGAACCGAAGACACTAGAATAACTAGT.............................................................................................. | 27 | 0 | 1 | 5.00 | 5 | 1 | 0 | 0 | 2 | 1 | 1 | 0 | 0 |
| .........................................................CGAAGACACTAGAATAACTAGT.............................................................................................. | 22 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTTGAACCGAAGACACTAGAATAAC.................................................................................................. | 26 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| ...........................................................................TAGTTACAAGTTATAGTTAACTAGT......................................................................... | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...........................................TATATCTGTTGAACCGAAGACACTAGAA...................................................................................................... | 28 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................CGAAGACACTAGAATAACTAGTT............................................................................................. | 23 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGTTGAACCGAAGACACTAGAAT..................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................CGAAGACACTAGAATTACTAGT.............................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................TTATATATCTGTTGAACCGAAGACA............................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TTTATATATCTGTTGAACCGAAGACAC........................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................TATTTATATATCTGTTGAACCGAA................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTGAACCGAAGACACTAGAATAACTAGT.............................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AGAACCGAAGACACTAGAATAACTAGT.............................................................................................. | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TTTATATATCTGTTGAACCGAAGA.............................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..TTGTCTGCACTAATAAGTTTTTCCCT................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................TTTATATATCTGTTGAACCGAATACAC........................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................TAGTGTCTTTGTTTGAACCACATCGTT......................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................TATATATCTGTTGAACCGAAGACACTAGA....................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................TGTTCTAGTGTCTTTGTTTGAACCACA.............................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGTCTCTTTGTTCGTACCA................................................ | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................GTTCGAGTCTCTTTGTT....................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......TTGCACTAATAACTTGTTCC................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
|
ACACTGAATAAAACTCATTTTGCAGAATGAAAAAAAAAGAAAACGATGTGGTTCAAACAAAGACACTAGAACAACTAGTTAACTATAACTTGTAACTAGTTATTCTAGTGTCTTCGGTTCAACAGATATATAAATATTTGTATCAAGGGAAAAACTTATTAGTGCAGACAAAT
***********************************...............(((.((.(((((((((((((((((((((((.((........))))))))))))))))))))))))).)).)))...............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553485 Ovary |
M025 embryo |
M053 Female-body |
SRR553487 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................TATTCTAGTGTCTTCGGTTCAACAGA............................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................CAAAGACACTAGAACAACTAGT.............................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................TGAAAAAAAAAGAAAACGATGTGGT......................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................AACAAAGACACTAGAACAACT................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................TGCAGAATGAAAAAAAAAGAAAACGA............................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................TCAAACAAAGACACTAGAACAACTAGTT............................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................TAGTTATTCTAGTGTCTTTGGTTCAAC.................................................. | 27 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TTCAACCAAAGACACTAGAAC..................................................................................................... | 21 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................AACAAAGACACTAGAAAAACT................................................................................................. | 21 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................TTGCAGAATGTAAAAAAAAA...................................................................................................................................... | 20 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | chrXh_Mrandom_005:3730-3902 + | dsi-mir-2581 | ATT-TGTCTGCACTAATAAGTTTTTCCCTTGATACAAATATTTATATATCTGTTGAACCGAAGACACTAGAATAACTAGTTACA---------AGTTATAGTTAACTAGTTGTTCTAGTGTCTTTGTTTGAACCACATCGTTTTCTTTTTTTTTCATTCTGCAAAATGAGTTTTATTCAGTGT |
| droSec2 | scaffold_109:22174-22351 + | ----TGTCTGCAATAATAAGTTTTTCCATTGATACAAATATTTATATATCTTTTGAACCAAAGACAGTAGAATTACTAGTTACTAGTTATACTAGTTATAGTTAACTAGTTATTCTAGTGTCTTTGTTTGAACCATATCGTTTT--TTTTTTTTCATTCTAAAAAATGAGTTTTATTCAGTGT | |
| dm3 | chrXHet:74898-74997 - | ATTTTGTCTGCAACGATAAGTTTTTCCCTTGATACAAATATTTATATATCTTTTGAGC---------------------------------------------------------------------------CATAGCGT--------TTTTTCATTCTGCAAAATTAGTTTTATTCAATGT | |
| droEre2 | scaffold_4845:20881752-20881836 + | CA-------------------------------------------------------------------------------------------GGTAACAGGTATCTGGTAGTCCGGGAA-------CTCGACCATAGCATTCTATCTTGTTTTCATTCTGCCAATAGCAATTTAATCAGTGT | |
| droYak3 | v2_chrUn_1329:26346-26446 + | AAT-TATCTGCAACGATAAGTTTTTCCTCTGTTACAAATATTTTAATATCTTTTGAACCAAA---------------------------------------------------------------------------GCGT------TTTTTTTCATTCTGCAAAATGAGTTTTATTCAGCGT | |
| droBip1 | scf7180000395874:7173-7286 + | ACG-CGACTGCAATATTAAATTTGCACATT-----AAATATTTGCATATTAATT-TGCAATATATATTAAAATAAAAAGCTAA------------------------------------------------------ACAA--------TATTACATTTTTTGAAATGAATTCCAATCTGTGT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 12/06/2013 at 02:41 PM