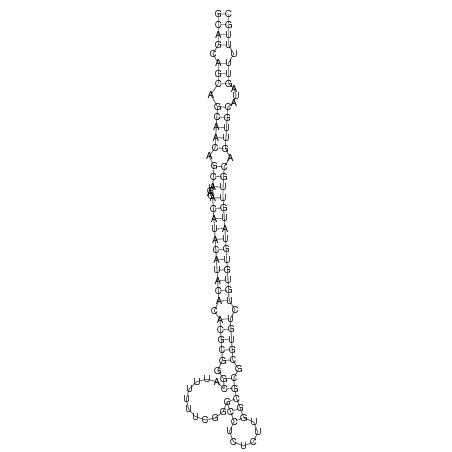
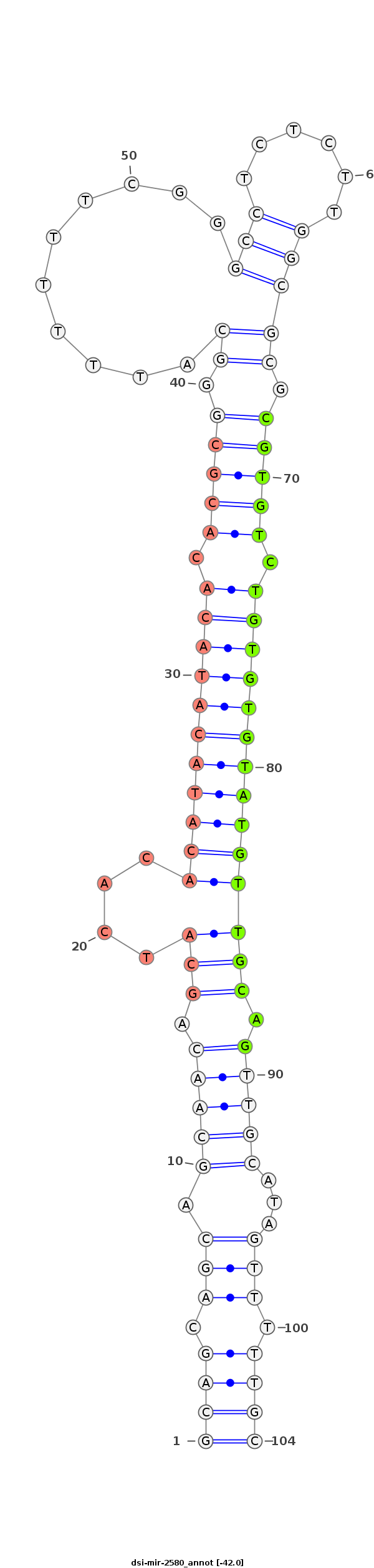
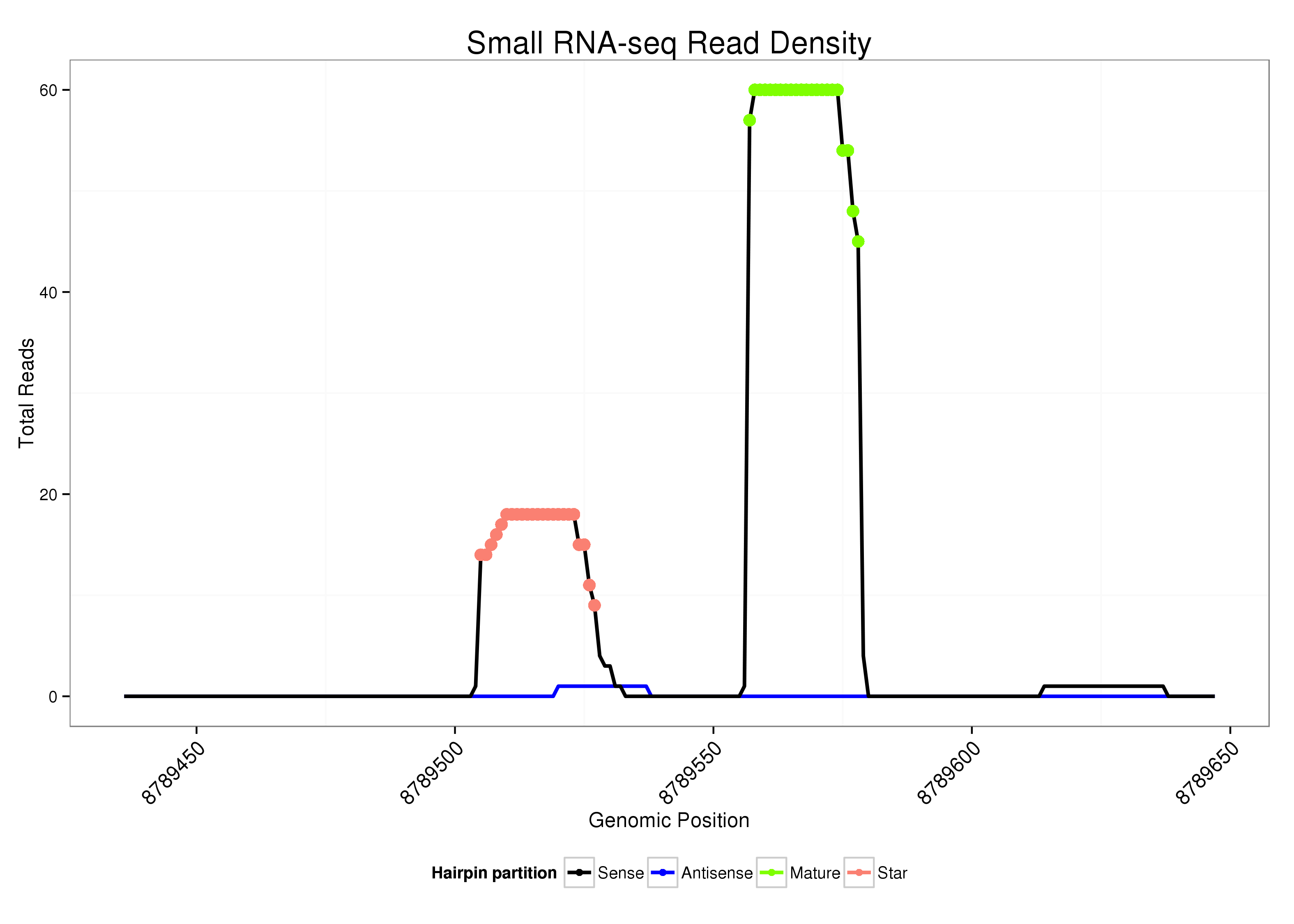
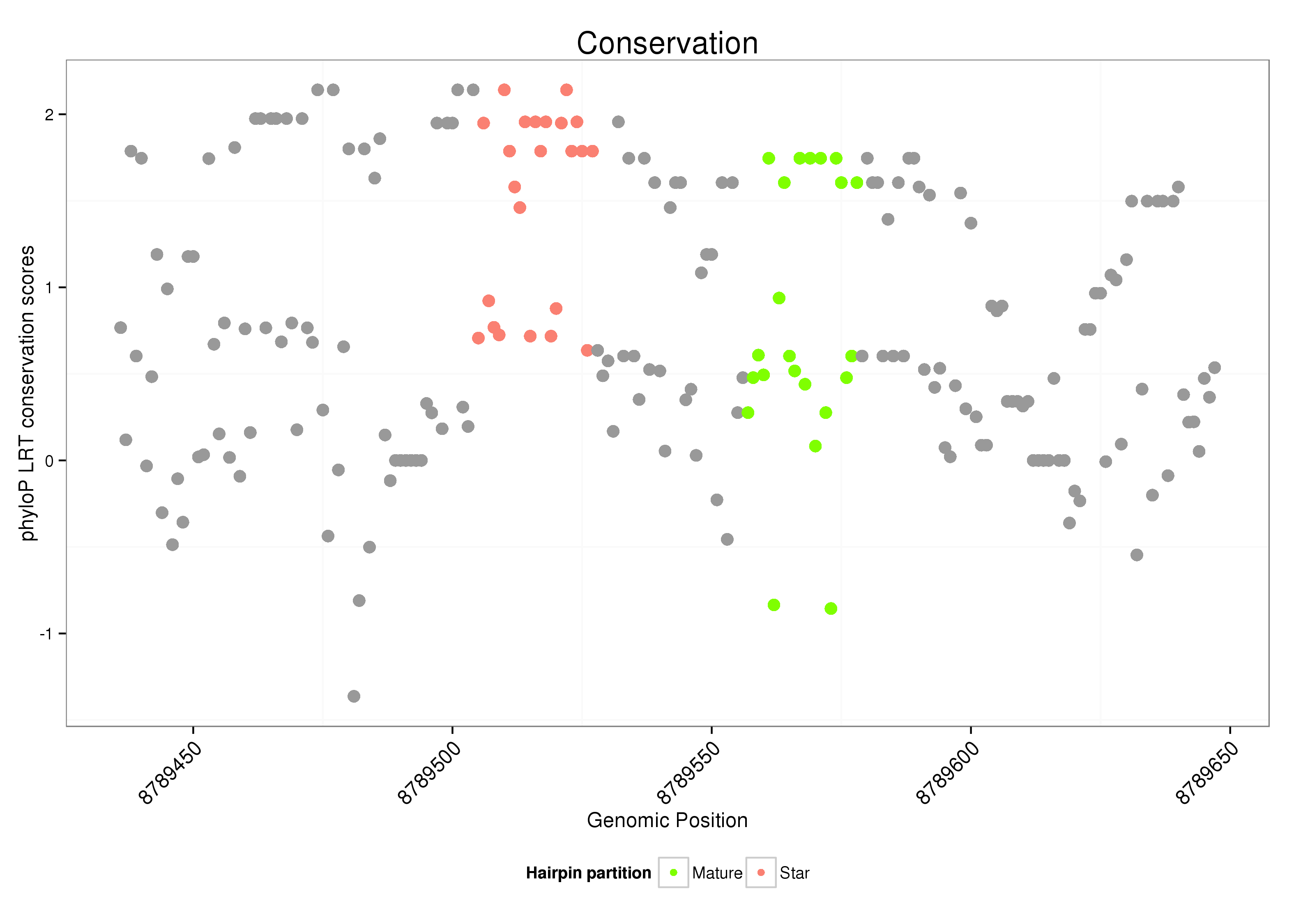
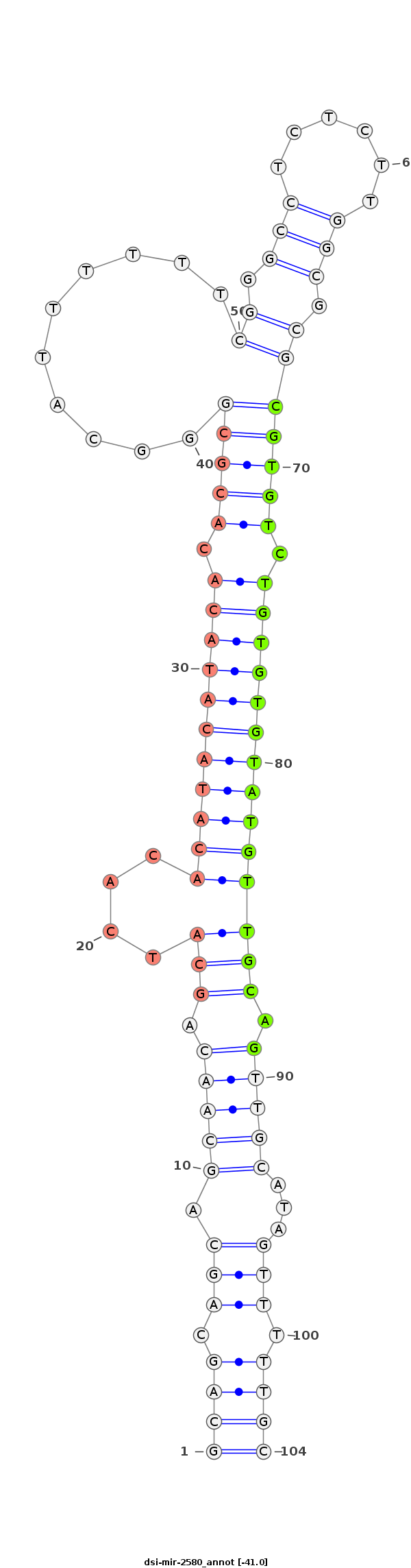
| .........................................................................................................................CGTGTCTGTGTGTATGTTGCAG..................................................................... |
22 |
0 |
1 |
39.00 |
39 |
8 |
5 |
6 |
2 |
3 |
3 |
5 |
0 |
2 |
4 |
1 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTATGTTGC....................................................................... |
20 |
0 |
1 |
6.00 |
6 |
3 |
0 |
0 |
0 |
1 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTATGTT......................................................................... |
18 |
0 |
1 |
6.00 |
6 |
2 |
0 |
0 |
0 |
0 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
| .....................................................................GCATCACACATACATACACACGC........................................................................................................................ |
23 |
0 |
1 |
4.00 |
4 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................GCATCACACATACATACAC............................................................................................................................ |
19 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTATGTTGCA...................................................................... |
21 |
0 |
1 |
3.00 |
3 |
0 |
0 |
1 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................GCATCACACATACATACACAC.......................................................................................................................... |
21 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................GCATCACACATACATACACACG......................................................................................................................... |
22 |
0 |
1 |
2.00 |
2 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| .........................................................................................................................CGTGTCTGTGTGTATGTTGCAGT.................................................................... |
23 |
0 |
1 |
2.00 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTATGTTGCAGA.................................................................... |
23 |
1 |
1 |
2.00 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................GTGTCTGTGTGTATGTTGCAGT.................................................................... |
22 |
0 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................GTGTCTGTGTGTATGTTGCAG..................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................GTGTCTGTGTGTGTGTTGCAG..................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .........................................................................................................................CCTGTCTGTGTGTATGTTGCA...................................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................GCATCACACATACATACACACGCG....................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................TTTTCGGGCCTCTCTTGAC.............................................................................................. |
19 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTATGTTGCAA..................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................GATTTTTTTTTTGTTTTTGCTGTT.......... |
24 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTGTGTTGCAGA.................................................................... |
23 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCGTGTCTGTGTGTATGTTGCAG..................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................TCACACATACATACACACGCGGG..................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................CAACAGCATCACACATACATACACACGA........................................................................................................................ |
28 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTGTGTTGCAG..................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .......................................................................ATCACACATACATACACACGCGGGCA................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................AGCATCACACATACATACACAC.......................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| .........................................................................CACACATACATACACACGCGGG..................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTGTGTTGCA...................................................................... |
21 |
1 |
2 |
1.00 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
| ..........................................................................ACACATACATACACACGC........................................................................................................................ |
18 |
0 |
4 |
1.00 |
4 |
0 |
0 |
0 |
0 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................TGTCTCTGTGTATGTTGCAGTT................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................CCTCTCTTGGCGCGA.......................................................................................... |
15 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................................................................TTGTTGCAGATGGATAGTTTT......................................................... |
21 |
3 |
4 |
0.50 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
| ......TTAACTGGCGAACTAC.............................................................................................................................................................................................. |
16 |
1 |
3 |
0.33 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGTCTGTGTGTAT............................................................................ |
15 |
0 |
4 |
0.25 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................TGTATCTTGCAGTAGCATGGT............................................................ |
21 |
3 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................CGTGGCTGTGTGTGTGTTGCAGA.................................................................... |
23 |
3 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................CCCACGCGGGCTTTTTTT............................................................................................................. |
18 |
2 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..............................................AACCAGCAGCAGCAGCA..................................................................................................................................................... |
17 |
0 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................GCGTGGCTGTGTGTATGTTTA....................................................................... |
21 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |