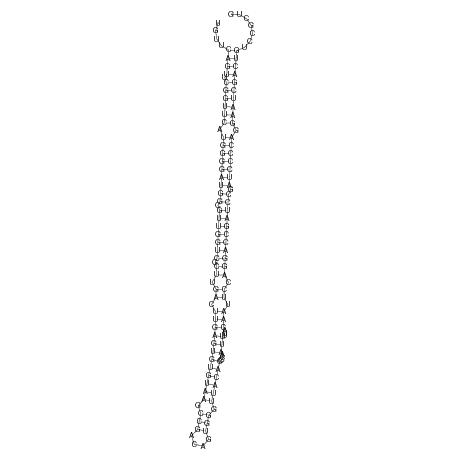
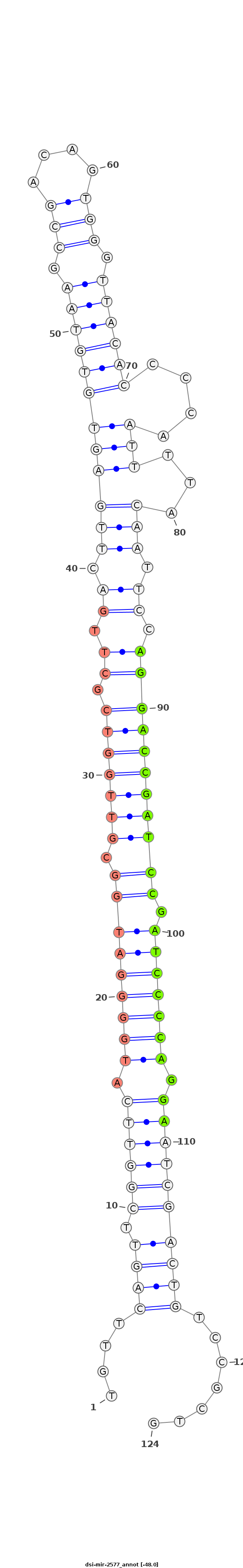
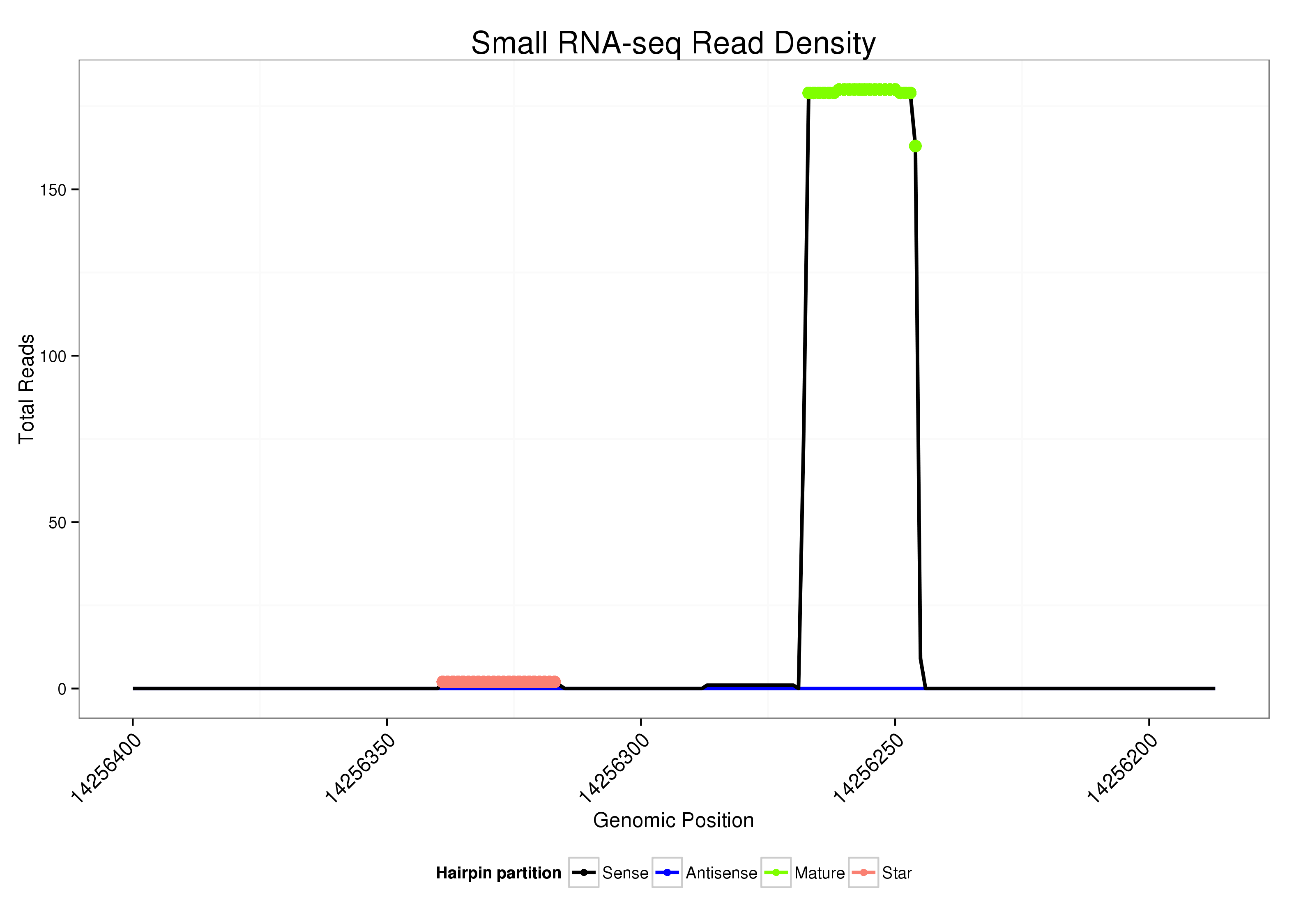
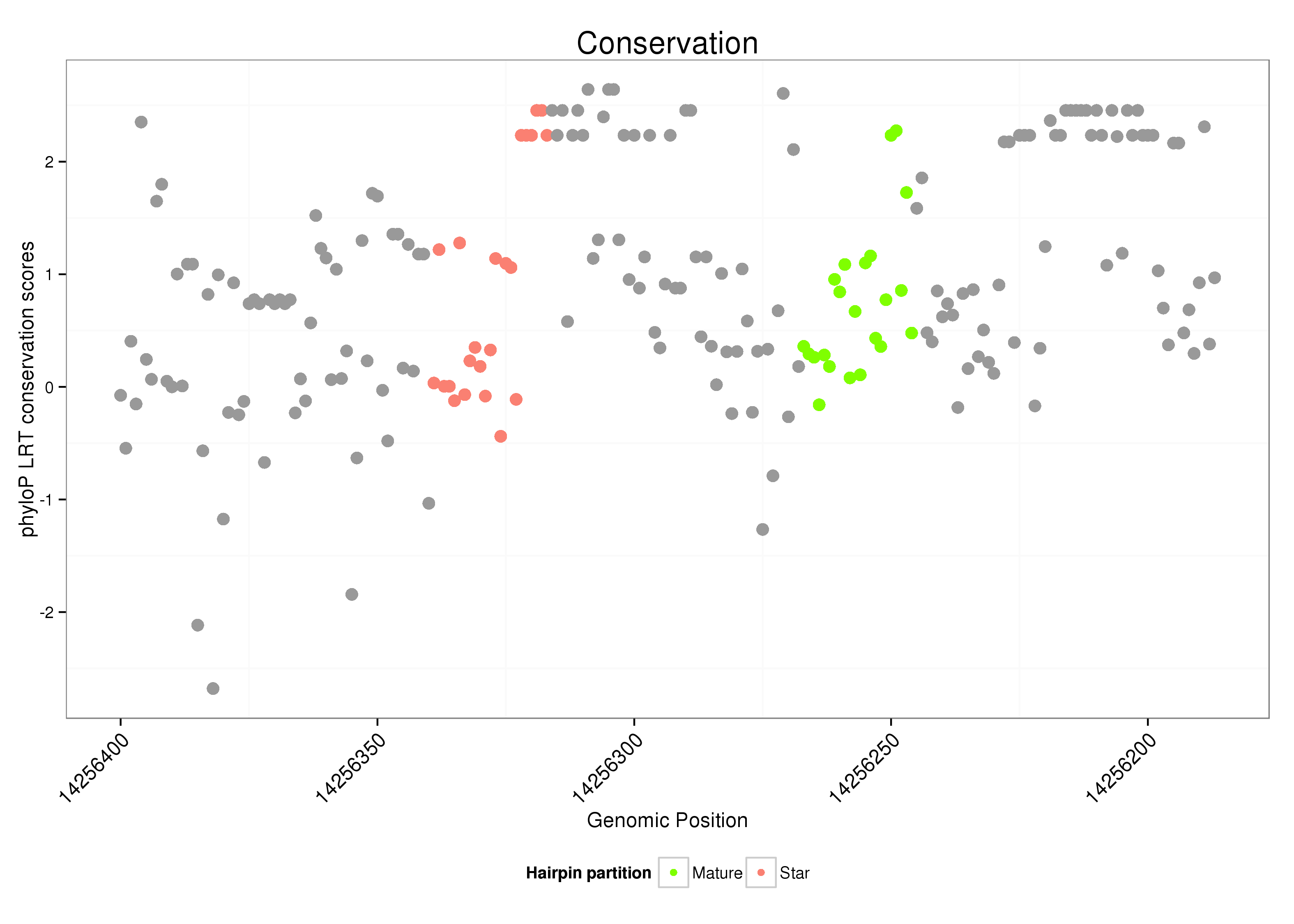
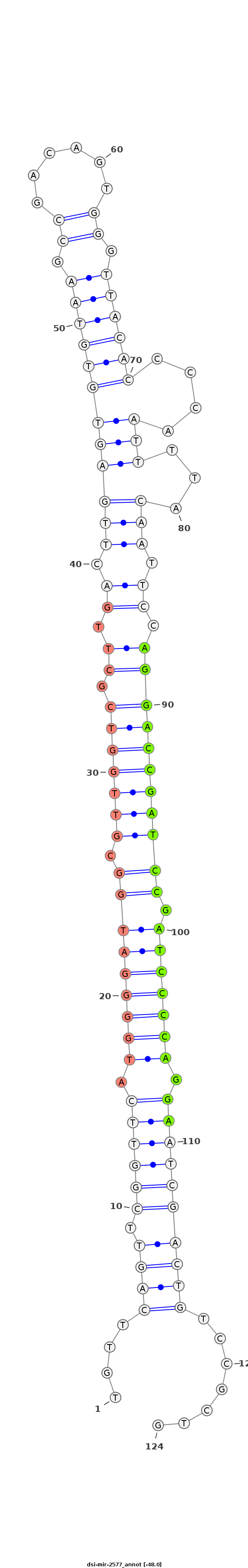
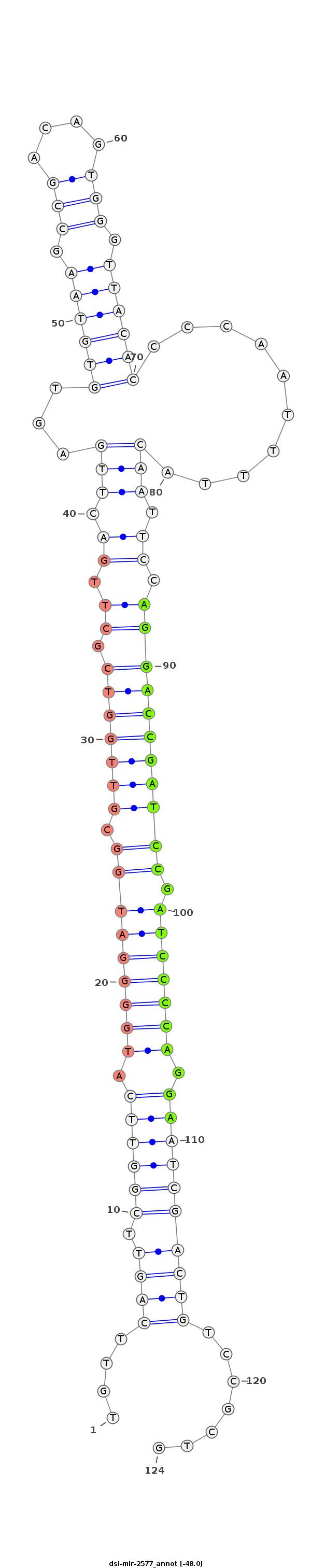
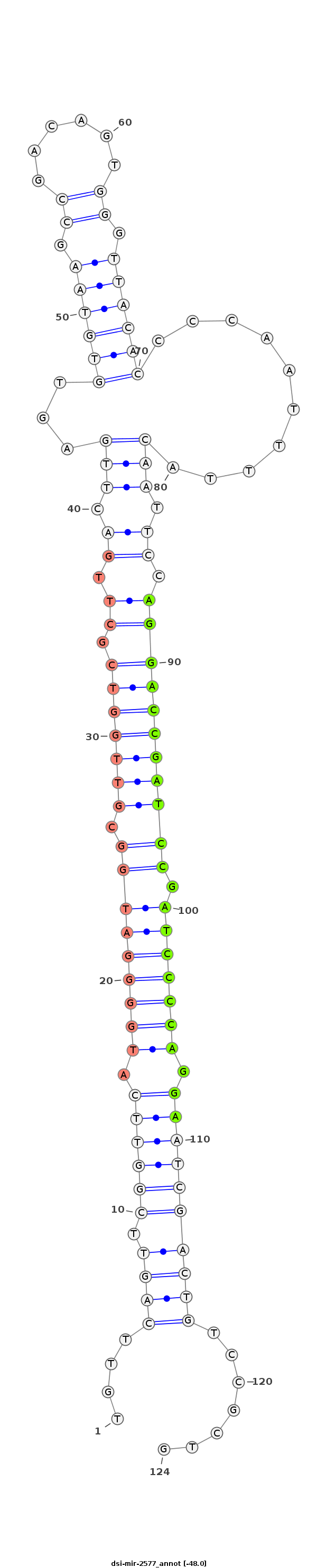
ID:dsi-mir-2577 |
Coordinate:2L:14256237-14256350 - |
Confidence:Known |
Type:Unknown |
[View on miRBase] [View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
| -48.0 | -48.0 | -48.0 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CAGTTCAGTTCGGTTGGGTTGAGCTGAGCTGAGTTGAGTTCAGTTCTGTTCAGTTCGGTTCATGGGGATGGCGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTTTACAATTCCAGGACCGATCCGATCCCCAGGAATCGACTGTCCGCTGTCGGGCGGACTGCCAATTACACTTTTTGACGCCGAGCACAATGT
**********************************************....((((.((((((.(((((((((.(((((((.((.((.((((((((((((.(((....))).))))))....)))...))).)).))))))))))).))))))).)))))))))).......******************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
GSM343915 embryo |
M023 head |
SRR553487 Ovary |
M024 male body |
O001 Testis |
SRR553488 Ovary |
SRR618934 Ovary |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................AGGACCGATCCGATCCCCAGGA........................................................... | 22 | 0 | 1 | 87.00 | 87 | 65 | 16 | 6 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCCAGGA........................................................... | 23 | 0 | 1 | 67.00 | 67 | 51 | 2 | 13 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCCAGGG........................................................... | 23 | 1 | 1 | 13.00 | 13 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCGATCCCCAGGG........................................................... | 22 | 1 | 1 | 11.00 | 11 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCGATCCCCAGG............................................................ | 21 | 0 | 1 | 10.00 | 10 | 1 | 6 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCGATCCCCAGGAA.......................................................... | 23 | 0 | 1 | 7.00 | 7 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCCAGG............................................................ | 22 | 0 | 1 | 5.00 | 5 | 3 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCCAGGAA.......................................................... | 24 | 0 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CGGGACCGATCCGATCCCCAGGA........................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CGGGACCGATCCGATCCCCAGG............................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GATCCGATCCCCAGG............................................................ | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................AGGACCGATCCGACCCCCAGGA........................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCC............................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCTATCCCCAGG............................................................ | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCGATCCCCCGG............................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................ATGGGGATGGCGTTGGTCGCTTG.................................................................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGAACGATCCGATCCCCAGGA........................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................ATGGGGATGGCGTTGGTTGCTTG.................................................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CACCCCAATTTTACAATT................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................CAGGACCGATCCGATCCCCAGGC........................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCAATCCCCAGGA........................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................ATGGGGATGGCGTTGGTCGCTTGA................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................ATGGGGATGGCGTTGGTCGATT................................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................AGGACCGATCCGATCCCCGTGG........................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TAGACCGATCCGATCCCCGGGA........................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................TTCTGTTCGGTTCATG...................................................................................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................GCCTTGGTCGCTTGCCTT.............................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CCCGCGAATCGACTGTCCGG.............................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TACAATTCCCGGACC........................................................................... | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................GAGTGAGTACTCCGACAGT........................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TGTGTAGGCCGACATTCGGT....................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................GGCGTTGGTTGCTTGTC................................................................................................................................ | 17 | 2 | 15 | 0.13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| ..............................................................TGGGGATAGCGTCGGTCCC..................................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TGGTGTTGGTCGCTTGGCT............................................................................................................................... | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................GGCGTTGGTTGCTTGCCT............................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................ACGTTGGTCGTTTGACTGGA............................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................GGCCTTGGTCGCTTGCC................................................................................................................................ | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TTTTACGATTATAGGACCG.......................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................ATCCCAAGAATCGACTGTC................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACATTGTGCTCGGCGTCAAAAAGTGTAATTGGCAGTCCGCCCGACAGCGGACAGTCGATTCCTGGGGATCGGATCGGTCCTGGAATTGTAAAATTGGGGTGTAACCCACTGTCGGCTTACACACTCAAGTCAAGCGACCAACGCCATCCCCATGAACCGAACTGAACAGAACTGAACTCAACTCAGCTCAGCTCAACCCAACCGAACTGAACTG
********************************************.......((((((((((.(((((((.(((((((((((.((.(((...(((....((((((.(((....))).)))))))))))).)).)).))))))).))))))))).)))))).))))....********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total |
|---|---|---|---|---|---|
| ....................................................................................................................................................CCCAGGAACCGAGCTGAAC............................................... | 19 | 2 | 2 | 0.50 | 1 |
| .................................AGCCCGTCCGACAGCG..................................................................................................................................................................... | 16 | 2 | 14 | 0.14 | 2 |
| ....................................................................................................................TTACACATTCAAGTCGAG................................................................................ | 18 | 2 | 8 | 0.13 | 1 |
| ............................................................................................................CTGTCGGAGTACTCACTCA....................................................................................... | 19 | 3 | 14 | 0.07 | 1 |
| ....TGTGATCGGCATCAAAAA................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 |
| .............................................................................TCCTGGAATTTTAAA.......................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 |
| ...................................................................................AATAGTAAAATTGGG.................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim3 | 2L:14256187-14256400 - | dsi-mir-2577 | C-AGTTCAGTTCGGTTGGGTTG---AG---------------------------------------------------------------------------------------CTGAGCTGAGTT----------------------------------------------------------------------------------GA-------GTTCA------------------------------------GTTCTGTTCAGTTCGGTT-CA-------------TGG-GGATGGCGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTT---------------------------------------------------------------------------------TACAATTCCAGGACCGATC-----------CGATCCCCAGGAATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGA-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT |
| droSec2 | scaffold_3:771865-772073 - | C-AGTTC-----GGTTGGGTTG---AGC---------------------------------------------------------------------------------------TGAGCTGAGTT----------------------------------------------------------------------------------GA-------GTTCA------------------------------------GTTCTGTTCAGTTCGGTT-GA-------------TGG-GGATGGCGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTT---------------------------------------------------------------------------------TACAATTCCAGGACCGATC-----------CGATCCCCAGGAATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGA-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| dm3 | chr2L:14508658-14508866 - | C-AGTTCAGTTCGGTTGGGTTG---AG---------------------------------------------------------------------------------------CTGAGCTGAGTT----------------------------------------------------------------------------------GA-------GTTGA------------------------------------G-----TTCAGTTCGGTT-GA-------------TGG-GGATGGCGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTT---------------------------------------------------------------------------------TACAATTCCAGGACCGATC-----------CGATCCCCAGGAATCGA-------------------------------------------------------------CTGTCCA-CT-GTCGGGCGGA-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| droEre2 | scaffold_4929:6262855-6263092 - | C-AGTTCAGTT-----CAGTTC---AGTTCAGTTC-------------------------------------------------------------------GGTTCGGTT-------------------------------------------------------------------------------------------------CG-------GTTCAGCTGGGTTGGATTG------GCTTGGACTGGGTTGAGCGGAGTTCA-------------------GCTGTTGG-GGATGGCGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTT---------------------------------------------------------------------------------TACAATTCCAGGACCGATC-----------CGATGC-CAGGCATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGA-CTGCCAATTACACATTTTGACGCCGAGCA-CAATGT | |
| droYak3 | 2L:14617624-14617872 - | C-AGTTCAGTT-----CAGCTT---GGTTTGGTTG-------------------------------------------------------------------GGTTGAG-----CTGAGTTGAGTT----------------------------------------------------------------------------------GA-------GT--------------------------TGATCTGAGTTCAGTTCAGTTCAGTTAGGTTATGTTGGTGAGGCTGATGGGGGATGGCGTTGGTCGCTTGACTCGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTT---------------------------------------------------------------------------------TACAATTCCAGGACCGATG-----------CGATCCCCAGGAATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGA-CTGCCAATTACACATTTTGACGCCGAGCA-CAATGT | |
| droEug1 | scf7180000409452:1074006-1074173 - | C-AGTTGAGTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CA------------------------------GTTCGGTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTCCA----------------CAATTCCA---------------------------------------------------------CATTTCCAGGTCCGATC-----------AGAGCCACAGGAATCGA-------------------------------------------------------------CTGTCCGCCT-GTCGGGCGGG-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| droBia1 | scf7180000302408:2046056-2046220 + | C-TGTTC-----GCTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCGGCCGCTTGACTCGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTCAA----------------CAATTCCA---------------------------------------------------------CAATGCCAGGACCGATC-----------CGATCCACAGGAATCGA------------------------------------------CTGTCGACCGGC-G----ACTGTCCG-CT-GTCGGACGGA-CTGCCAATTACACTTTTTGACGCT------------ | |
| droTak1 | scf7180000415399:736271-736446 + | C-G------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTTGGTCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTCCA----------------CAGTTCTA---------------------------------CAGTTCTACGTTTCCACAGCTCTACAGTTCCAGGACCGATC-----------CGATCCACAGGAATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGA-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| droEle1 | scf7180000491046:942103-942254 - | T----TCAGTT-----CAGATAGAGAGA-GAGCCGGG------------------------------------------------------------------------------------------------------------------------------------T------------------------------------------------------------------------------------------------------------------------------------------CCGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCGATT-----------------------------------------------------------------------------------GCGATTCCAGGTCCG---------------GATCCACAGGAATCGA-------------------------------------------------------------CTGA----CT-GT----CGGG-CTGCCAATTACACTTTTTGACGCCGAGCA-CGATG- | |
| droRho1 | scf7180000780078:27625-27864 - | C-AG---------------------AGA-GAGCCGGGTCCGGACTTGGGTTTTGGTATGGGTTTTGGTATGAGTTTGGGTATGAGTTTTGGGC-------------------------------------------------------------------------------TT----------------------------------GG-------GT----------------------------------------------TCG--------------------------------TCGGTCAGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCGATTCCGATTC------------CAATTCCG---------------------------------AA--------------------TCCGATTCCAGGGCCG---------------GATCCACAGGAATCGA---------------------------------------------C----CGGCTG----ACTGTCCG-CT-GTCGGGCGGG-CTGCCAATTACACTTTTTGACGCCGAGCA-CA---- | |
| droFic1 | scf7180000453843:263124-263331 + | C-AGA-------------GAGA---GAA-GAGCCGGG------------------------------------------------------------------------------------------------------------------------------------TTTTCTGTCGGTCGGCTGGTCGT-----------T---TGGTCGGTGGG-TCG--------------TTTGTTGGTT---------------------------------------------------GG-TCCGCAGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAATTC----------------------------------------------------------------------------------GCAATTCCAGGACC----------------GATCCACAGGAATCGA-------------------------------------------------------------CTGTCCG-CT-GTCGGGCGGG-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATAC | |
| droKik1 | scf7180000302473:885966-886147 - | CCAGCTC-----GCTTGAGCTC---AGA-GAGCCGG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCGCTCGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCGATTTCGAGTC------------CTGTTCCAAGTCCGACTC-----------------------------------------------CGAT-----------TCCGATCCCGAACCAATCCACAG-----------------------------------------------------------------------TC----G-GGCGGGCGGG-CTGCCAATTACACTTTCTGACGCCGAGCA-CAATGT | |
| droAna3 | scaffold_12916:1310203-1310426 + | C-AGCTC-----GCTTGAGTTC---AGA-GAGCCGGG------------------------------------------------------------------------------------------------------------------------------------TTCTC----------------------------------GG-------G---------------------------------------------------------------------------------TTCGGTTGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACCCCAGTTCC-----------------CT------------GCTCCGATCCCGTCCTCGTTCCCATTTCCG-TTAACGTTTCC-----TTTCCCATCCCTGATCC----------CAATTCAATCCACAGG-----A-------------------------------------------------------------CTGTCCG-CC-ATCGGACGGT-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| droBip1 | scf7180000396554:946575-946795 - | C-AGCTC-----GCTTGAGTTC---AGA-GAGCCGGG------------------------------------------------------------------------------------------------------------------------------------TTCTC----------------------------------GG-------G---------------------------------------------------------------------------------TTCGGTTGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTACACACCAGTTTGCCG--GCTTCCTGCCCCCT------------GCTCCG------------------TTTACG-TTTACGTTTCC-----TTTCCCATTCCTGATCC----------CAATTCAATCCACAGG-----A-------------------------------------------------------------CTGCCCG-CC-ATCGGGCGGT-CTGCCAATTACACTTTTTGACGCCGAGCA-CAATGT | |
| dp5 | 4_group1:3353981-3354211 + | T-TG---GGTT-----TGGGT-----------------------------------------------------------------TTTGGGTTGAGGTTCAGGTTCGCTTCAGTAGAGCTGAGTGGAGAGTTGCTGTGGTGCGGTGCGGCGGTGGAGTGAAGTGGAAGTTT---------------------------------------------------------------------------------------------TCG------------------------------GTTCGGTTGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTATATTCGTACTT-----------------------------------------------------------------------------------GTATTCGATAAAG----------------GATCC-C------------------------------------------------------------------------GGCAA-CT-GACGGGCGGGTTTGCCAATTACACTTTTTGACGCCGTGCGGCAGTG- | |
| droPer2 | scaffold_5:1869194-1869391 - | C-GCTTCAGTA-----GAGATG---AGTGGAG-------------------------------------------------------------------------------------------------AGTTGCTGTGGTGCGCTGTGGCGGTGGAGTGAAGTGGAAGTTT---------------------------------------------------------------------------------------------TCG------------------------------GTTCGGTTGGACGCTTGACTTGAGTGTGTAAGCCGACAGTGGGTTATATTCGTACTT-----------------------------------------------------------------------------------GTATTCGATAAAG----------------GATCC-C------------------------------------------------------------------------GGCAA-CT-GACGGGCGGGTTTGCCAATTACACTTTTTGACGCCGTGCGGCAGTG- | |
| droWil2 | scf2_1100000004577:2827268-2827508 + | T-ATCTGTGTG-----TCGGTC---TGTTGGT-------------------------------------------------------------------------------------------------------------------------------------------------------------CGGACTGACTGACTCTGACGG-------TT-----------------------------------------TGTGCTCGTTTCGTTT-CG-------------TTT-CGGTACGGTTGGACGCTTGACTTGAGTGTGTAAGACCGCCCCGCCTTGTGACTACCCTCCC----------------C------C---------------------------------AA--------------------GTTGATGCCAGGAAC---------------CGCTTTACAGGAATACA----------------------------GGAATATGGACACTCA-----------CATCCACATCGAG-ATCTACGGGCGGG-TTGCCAATTACACTTTTTGACGCCAGGCG-ACGTGT | |
| droVir3 | scaffold_10221:329-425 + | C-GGTTCAGTT-----CGATTC---GGTTCGGTTC-------------------------------------------------------------------AGTTCGGTT-------------------------------------------------------------------------------------------------CG-------GTTCA------------------------------------GTTTGGTTCAATTCGGTT-CG----C--GGCGAATGG-GGCATGCGTTGAT--------------------------------------------GTG----------------------------------------------------------------------------------CGAT----------------------------------------------------------------------------------------------------------------------------TGG-------------------------------CT | |
| droMoj3 | scaffold_6473:16527898-16527963 - | C-AGTTCAGTT-----CAGTTC---AGTTCAGCTC-------------------------------------------------------------------AGTTCAGTG------------------------------------------------------C------------------------------------------GA-------GTTCA------------------------------------GTTCGGTTCGGTTCGG------------------------------------------------TACGTAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15126:847690-847877 + | G-G--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGGACGCTTGACTTGAGTGTGTAAGCCGACGTCGGGTTACACACACCCCT------------------------------------------------------------------------------------CATCCGCAAACG---------------CAATCG-CAA---TCGGACTGGCAATGGCAATGGCAATGGGAATGGGAATGGGAATAGGCAAC----GGAGTGAGCGGGATTCGG-GT-TTAGGGCGGA-CTGCCAATTACACTT-TTGACGCCAGGCA-ACGTGT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 12/06/2013 at 02:37 PM