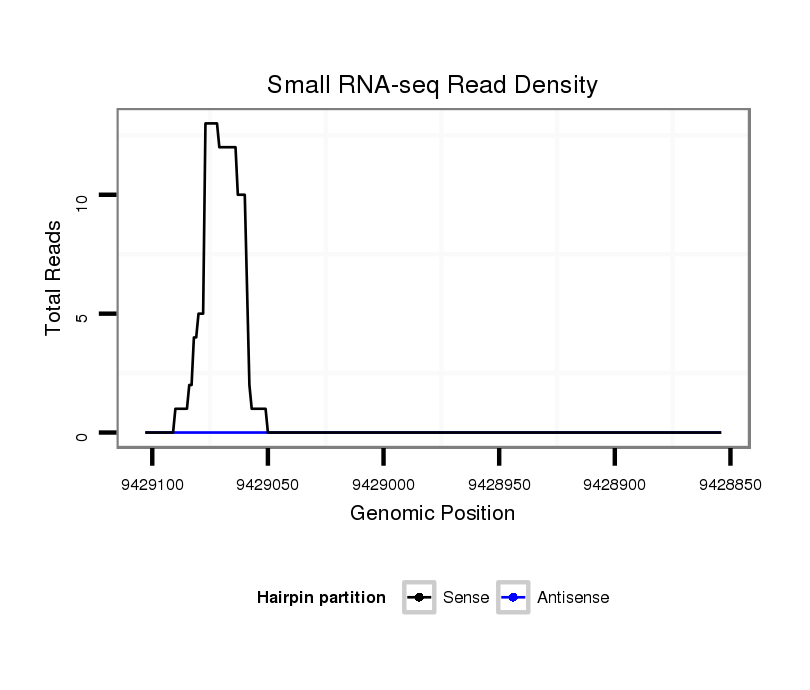
| GTTTTCGGGCGCTGTGCTGACAACATCC-TCGCAGGA-CCAAGGAGCGGATG-GTGCGTTCAA-TAGTCC----------------------T-------------------------CCT--------G--CTCG-------------------GTGTATGTGTGTGCAGAAACTTTTAACCGTGTGCGCTG-----------------TC-------------------------TGTGTGCGAGT----------GT--------TGG----TGCGCGAGGTGTTGGCAAATTGATTGCAGTTTGA---AATCAA---------------AGGGA-----------A-------------------AACTT-----------------------AAGCCAAATCGAA----------------------AAAGCT------------C-ACCT-----CAG-AATCGAAAGT---GCC------------CTAATCTGGGCGTACTCAAGGGG--------AGACA | Size | Hit Count | Total Norm | Total | M054
Female-body | V113
Male-body | V114
Embryo | V115
Head |
| ..........................CC-TCGCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 13.00 | 13 | 0 | 1 | 0 | 12 |
| ..........................CC-TCGCAGGA-CCAAGGAGCGGA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 4.00 | 4 | 0 | 1 | 0 | 3 |
| ...........................C-TCGCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 4.00 | 4 | 0 | 0 | 0 | 4 |
| ..........................CC-TCGCAGGA-CCAAGGAGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 3.00 | 3 | 1 | 0 | 0 | 2 |
| ..............................CGCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 3.00 | 3 | 0 | 0 | 0 | 3 |
| ..........................CC-TCGCAGGA-CCAAGGAG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 3.00 | 3 | 1 | 0 | 0 | 2 |
| ..............................CGCAGGA-CCAAGGAGCGGATG-GTGCG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ....................................A-CCAAGGAGCGGATG-GTGCGTTCAA-................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ...............................GCAGGA-CCAAGGAGCGGA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ...........................C-TCGCAGGA-CCAAGGAGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ...................................GA-CCAAGGAGCGGATG-GTGCG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ....................................A-CCAAGGAGCGGATG-GTGCG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ...............................GCAGGA-CCAAGGAGCGGATG-............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................ACCGTGTGCGCTG-----------------TC-------------------------TGTG................................................................................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ......................ACATCC-TCGCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................TCC-TCGCAGGA-CCAAGGAGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .................................AGGA-CCAAGGAGCGGATG-GTGCGT...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..............................CGCAGGA-CCAAGGAGCGGATG-GTG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..........................CC-TCGCAGGA-CCAAGGAGCGGATG-............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ........................ATCC-TCGCAGGA-CCAAGGAGC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..................................GGA-CCAAGGAGCGGATG-GTGCG....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ........................ATCC-TCGCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .................................AGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................GCGCTG-----------------TC-------------------------TGTGTGCGAGT----------G.............................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ...............................GCAGGA-CCAAGGAGCGGATG-G........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |