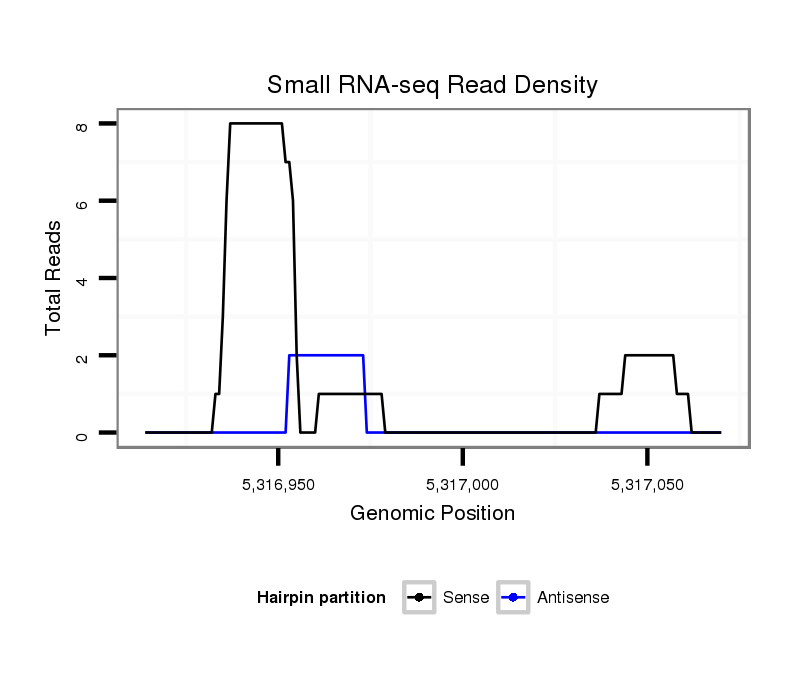
ID:dsi_9780 |
Coordinate:2r:5316964-5317020 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TCTCAAGCCCAGCTATGATCACTTCGATCTCAAGAGCTCCCGCAGTTACGGTAAGTCCAAAGAGAACTGAAAAACAGTAGAATAGTACAATCTAAATGTTGTCTTAGTTCGCAATACAGCCATCCTGAGCAACGACAGCGATGTGAACGCGGGGGAG
**************************************************.........((((.(((.........(((......))).........))).))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
M053 female body |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................TCGATCTCAAGAGCTCCC.................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TTCGATCTCAAGAGCTCCCGAAGTT.............................................................................................................. | 25 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TTCGATCTCAAGAGCTCCC.................................................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CTTCGATCTCAAGAGCTCCCG................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CTTCGATCTCAAGAGCTCC..................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GATCTCAAGAGCTCCCGCAGTTACGTTCG....................................................................................................... | 29 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................ACGGTAAGTCCAAAGAGA............................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................CACTTCGATCTCAAGAGCT....................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AACGACAGCGATGTGAAC......... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................TTCGATCTCAAGAGCTCCCG................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................CCTGAGCAACGACAGCGATGT............. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................AGTCCAAAGAGAACGG........................................................................................ | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................TTGTCTTAGTTCGGA............................................ | 15 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGTTACGGTCAGTCCAAA................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................TATAATCAAAATCTTGTCTTA................................................... | 21 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .............................................TAACTGTTAGTCCAAAGA.............................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................CAGTTGCGGTAATTCCA.................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AGAGTTCGGGTCGATACTAGTGAAGCTAGAGTTCTCGAGGGCGTCAATGCCATTCAGGTTTCTCTTGACTTTTTGTCATCTTATCATGTTAGATTTACAACAGAATCAAGCGTTATGTCGGTAGGACTCGTTGCTGTCGCTACACTTGCGCCCCCTC
**************************************************.........((((.(((.........(((......))).........))).))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M023 head |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................AGGGCTTCAATGCCAT........................................................................................................ | 16 | 1 | 1 | 6.00 | 6 | 3 | 2 | 0 | 0 | 1 | 0 | 0 |
| ...................................AGAGGGCTTCAATGCCAT........................................................................................................ | 18 | 2 | 1 | 3.00 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................GGCGTCAATGCCATTCAGGTT................................................................................................. | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ...................................AGAGGGCTTCAATGCCA......................................................................................................... | 17 | 2 | 4 | 1.25 | 5 | 2 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................AGGGCTTCAATGCCATT....................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TCGAGGGCTTCAATGC........................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................AGGGCTTCAATGCCATG....................................................................................................... | 17 | 2 | 11 | 0.55 | 6 | 2 | 3 | 0 | 0 | 1 | 0 | 0 |
| .....................................AGGGCTTCAATGCCA......................................................................................................... | 15 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................AGGGCTTCAATGCCATA....................................................................................................... | 17 | 2 | 6 | 0.33 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................GAGGGCTTCAATTCCAT........................................................................................................ | 17 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGAGGGCTTCAATGCCATA....................................................................................................... | 19 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................GAGGGCTTCAATGCC.......................................................................................................... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................GTCGGTAGGACTCGTTAGTT..................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................CCAATGCCATTCAGC................................................................................................... | 15 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AGAGGGCTTCAATGCCATG....................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................TATGTCGGCAGGGTTCGTT......................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................AGTGAAGCCCGAGTACTCG........................................................................................................................ | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................AGGGCTTCAATGCCAC........................................................................................................ | 16 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................AAGAGGGCTTCAATGCCAT........................................................................................................ | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................GGAGGGCTTCAATGC........................................................................................................... | 15 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................AATGCCATTCAGCTGCCT.............................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GTTTGTCTTGACTTTT.................................................................................... | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................AGAGGGCTTCAATGCCAC........................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................AGGGCTTCAATGCCACTA...................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/23/2015 at 12:58 PM