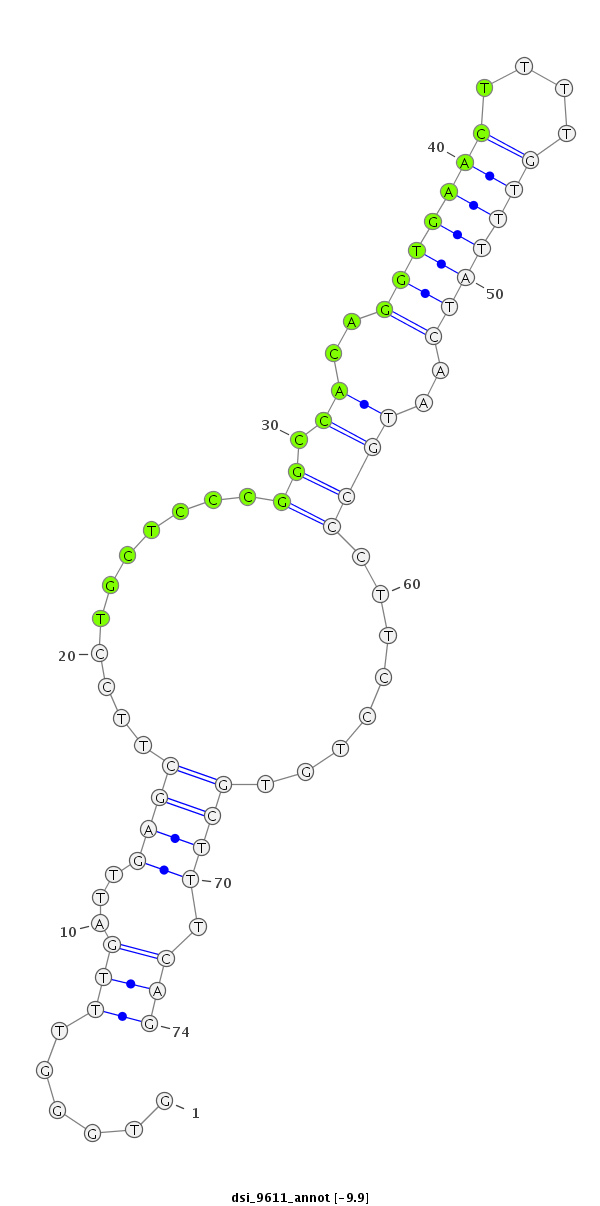
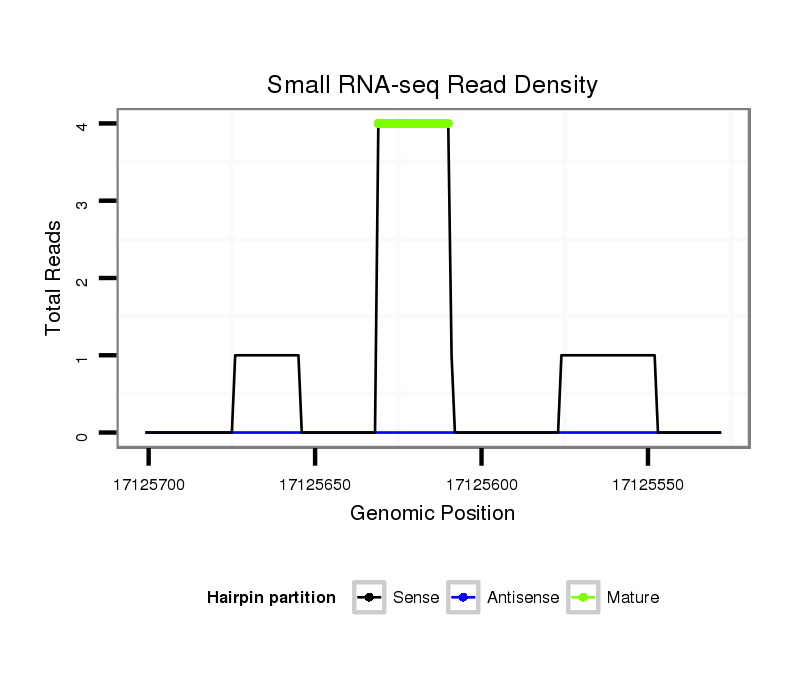

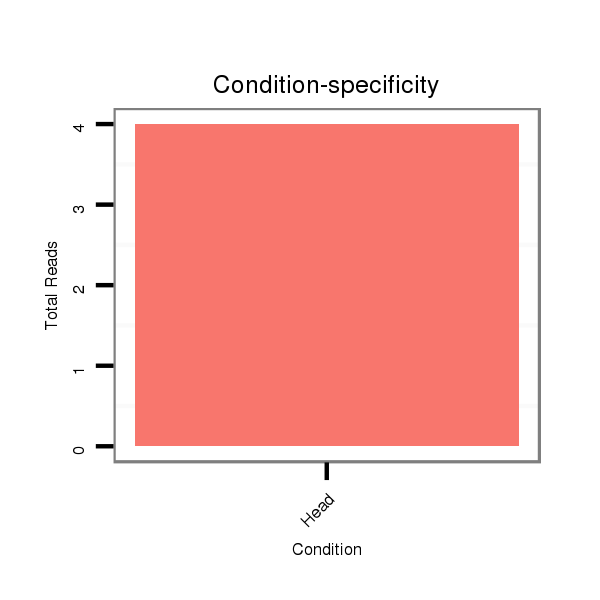
ID:dsi_9611 |
Coordinate:3r:17125578-17125651 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -16.7 | -16.6 | -15.9 | -15.7 |
 |
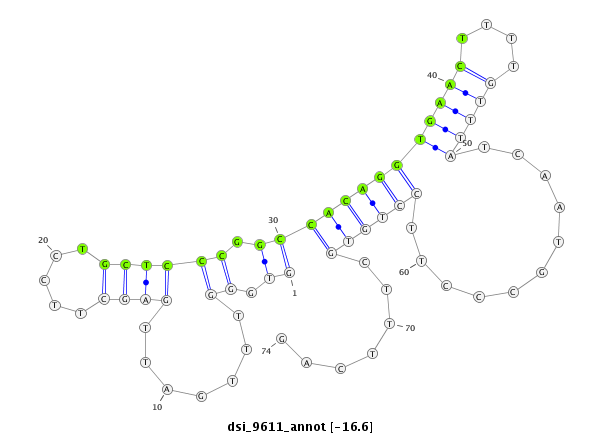 |
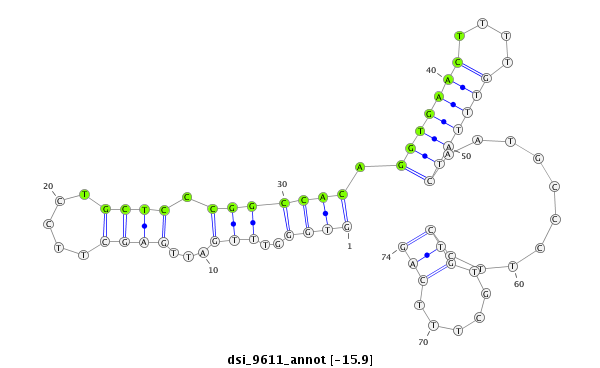 |
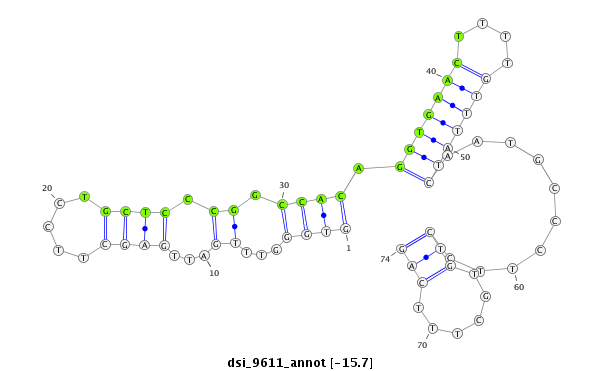 |
intergenic
No Repeatable elements found
|
CTTCGACACGCTCGATGTGGTCACGTACGAGGAGGTTGTCAAGGTGCCAGGTGGGTTTGATTGAGCTTCCTGCTCCCGGCCACAGGTGAACTTTTGTTTATCAATGCCCTTCCTGTGCTTTCAGTTGGCGACCCGAACTTTCAGCGCAAAACGTTGGTGCTCTTGGGTGCACAT
**************************************************......(((...((((...........((.((..(((((((....)))))))..))))........)))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
M053 female body |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TGCTCCCGGCCACAGGTGAACT.................................................................................. | 22 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................TGGCGACCCGAACTTTCAGCGCAAAACGT.................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................TGAACATTTGTTTATCAACGCC.................................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................CGAGGAGGTTGTCAAGGTGC............................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................TGCTCCCGGCCACAGGTGAACTT................................................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................GTGCATTCAGTTGGCGAC.......................................... | 18 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TGCTCCCGGCCACAGGTGAACC.................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CTGTGCGTCCCGTTGGCGAC.......................................... | 20 | 3 | 9 | 0.22 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................CGGGTGAACAGTTGTTTATC........................................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................TACCAGGTGGTTGTCA..................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................CTTCCCGCTCCCGGC.............................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................GAGGAGGTCGACAAGGTG................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GAAGCTGTGCGAGCTACACCAGTGCATGCTCCTCCAACAGTTCCACGGTCCACCCAAACTAACTCGAAGGACGAGGGCCGGTGTCCACTTGAAAACAAATAGTTACGGGAAGGACACGAAAGTCAACCGCTGGGCTTGAAAGTCGCGTTTTGCAACCACGAGAACCCACGTGTA
**************************************************......(((...((((...........((.((..(((((((....)))))))..))))........)))).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|
| .........................................................................AGGGCAGGTGTGCACT..................................................................................... | 16 | 2 | 20 | 0.40 | 8 | 8 |
| .........................................................................AGGGCAGGTGTGCACTT.................................................................................... | 17 | 2 | 5 | 0.40 | 2 | 2 |
| .........................................................................AGGGCAGGTGTGCACTTG................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 1 |
| ..................................................................................................................................TGGGGTTGTAAGTCGCGT.......................... | 18 | 2 | 5 | 0.20 | 1 | 1 |
| ..........................................................................GGGCAGGTGTGCACTT.................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 |
Generated: 04/23/2015 at 12:37 PM