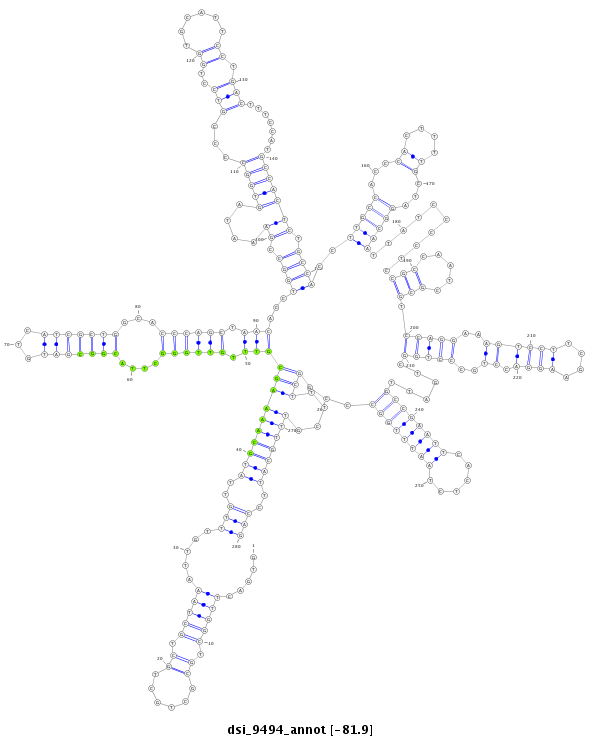
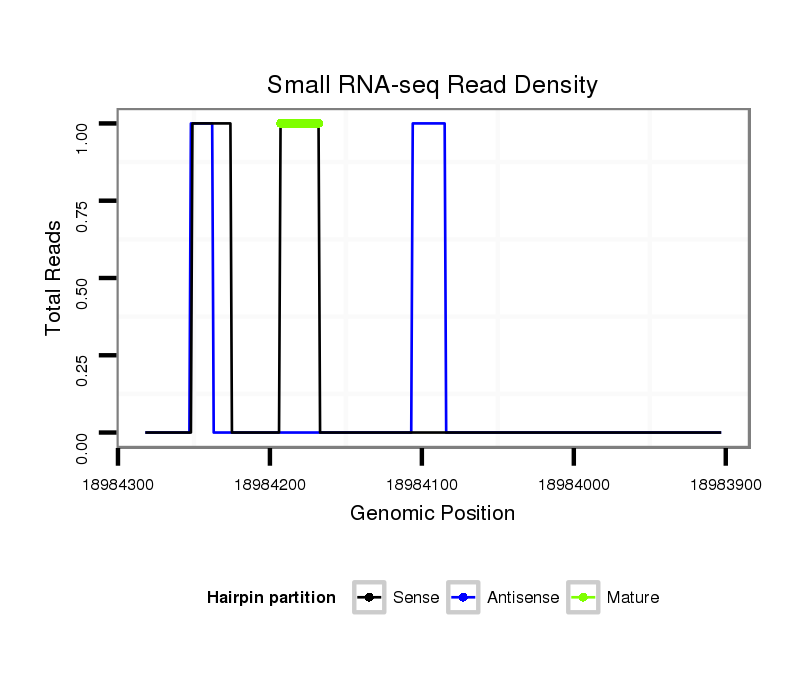
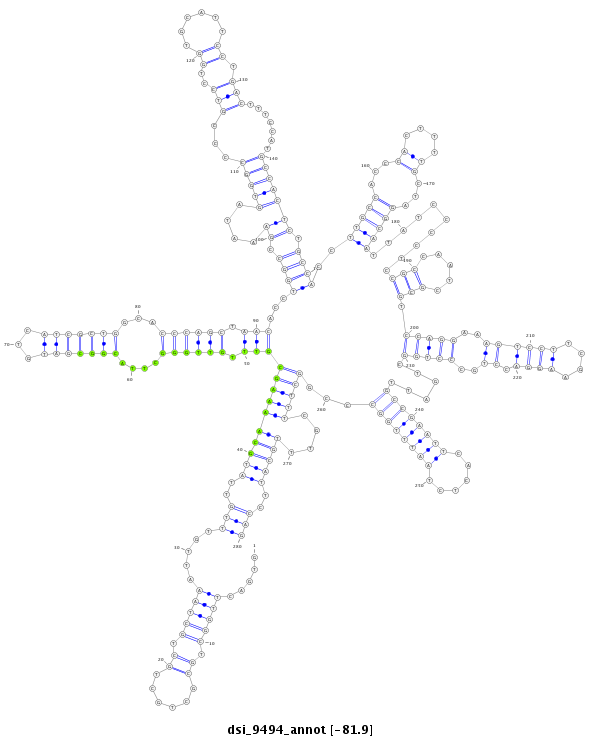
| ..........................CCAAGGAACCTCTGGTCGAC.............................................................................................................................................................................................................................................................................................................................................. |
20 |
2 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGTGAGCCGGTAGAACGG............................................................................................................................................................................ |
18 |
2 |
19 |
1.26 |
24 |
0 |
0 |
20 |
0 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................CCAAGGAACCTCTGGTC................................................................................................................................................................................................................................................................................................................................................. |
17 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................TCCCGGGAACCTCTGGTCGACG............................................................................................................................................................................................................................................................................................................................................. |
22 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................GGACTGAAAGGTACGGTGAGAC...................................................................................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................CCAAGGAACCTCTGGT.................................................................................................................................................................................................................................................................................................................................................. |
16 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................TAAGGACTGAAACGGACGTTGAG........................................................................................................................................................................................ |
23 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................CCAAGGAACCTCTGGTCGA............................................................................................................................................................................................................................................................................................................................................... |
19 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGTCCA............................................................................................................................................................................................................................................................................................................................................... |
15 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGACTACG............................................................................................................................................................................................................................................................................................................................................. |
17 |
2 |
11 |
0.73 |
8 |
0 |
0 |
3 |
4 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................CCAAGGAACCTCTGG................................................................................................................................................................................................................................................................................................................................................... |
15 |
1 |
8 |
0.63 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................CGCGGACGAGGAACCTCTGGT.................................................................................................................................................................................................................................................................................................................................................. |
21 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................................GTTAGTGTAGGTCCATTCAGG....................................................................................................................... |
21 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................AGGAACCTCTGGTCGACG............................................................................................................................................................................................................................................................................................................................................. |
18 |
1 |
5 |
0.40 |
2 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGACCATGG............................................................................................................................................................................................................................................................................................................................................ |
18 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....GCGGCTGTAGTAGCTA....................................................................................................................................................................................................................................................................................................................................................................... |
16 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........CGGTAGTAGCTACGAGGCGCAG............................................................................................................................................................................................................................................................................................................................................................. |
22 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................TGGAACCTCTGGTCC................................................................................................................................................................................................................................................................................................................................................ |
15 |
1 |
17 |
0.29 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................AACCTCTGGACCACG............................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
7 |
0.29 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGTCTA............................................................................................................................................................................................................................................................................................................................................... |
15 |
1 |
15 |
0.27 |
4 |
0 |
3 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................CCATGGAACCTCTGGTCGACG............................................................................................................................................................................................................................................................................................................................................. |
21 |
3 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGTGAGACGGTGGAGAGG............................................................................................................................................................................ |
18 |
2 |
20 |
0.25 |
5 |
1 |
0 |
0 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................GCGTCCGAGGAACCTCTGG................................................................................................................................................................................................................................................................................................................................................... |
19 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGTGAGCCGGTGGAACGG............................................................................................................................................................................ |
18 |
1 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................GAACCTCTGGTCTACG............................................................................................................................................................................................................................................................................................................................................. |
16 |
1 |
5 |
0.20 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................TGTGGAACGGCTTTGTCACG............................................................................................................................................................................................................................. |
20 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................AACCTCTGGTCCACGC............................................................................................................................................................................................................................................................................................................................................ |
16 |
1 |
5 |
0.20 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................................GTTAGTGTAGGTCCATTCAG........................................................................................................................ |
20 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................CCGGGAACCTCTGGTCGA............................................................................................................................................................................................................................................................................................................................................... |
18 |
2 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................TGTGGACCGGTTGTATCA............................................................................................................................................................................................................................... |
18 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......GGCTGTAGTGGCTAC...................................................................................................................................................................................................................................................................................................................................................................... |
15 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................AGGAACCTCTGGTCGACGA............................................................................................................................................................................................................................................................................................................................................ |
19 |
2 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ............................AAGGAACCTCTGGTCGACG............................................................................................................................................................................................................................................................................................................................................. |
19 |
2 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....GGCGGCTGTAGTGGCTAC...................................................................................................................................................................................................................................................................................................................................................................... |
18 |
2 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................................................................AGCTTCCGGAACGGGTCCGAC................................................................................................. |
21 |
3 |
9 |
0.11 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....GCGGCTGTAGTGGCTAC...................................................................................................................................................................................................................................................................................................................................................................... |
17 |
2 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................AGGACTGACAGATACGG............................................................................................................................................................................................ |
17 |
2 |
10 |
0.10 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................TGAGACGGTGGAAAGG............................................................................................................................................................................ |
16 |
1 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................................................GCGGCTAGCGCAGGT................................................................................................................................ |
15 |
1 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGTGAGACGGTGGAAAGA............................................................................................................................................................................ |
18 |
2 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ............................AAGGAACCTCTGGTCGAC.............................................................................................................................................................................................................................................................................................................................................. |
18 |
2 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................GTCGAGTGTGGACCG........................................................................................................................................................................................................................................ |
15 |
1 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGTCGAAGGTA.......................................................................................................................................................................................................................................................................................................................................... |
20 |
3 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................AACCTCTGGTCTACG............................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
14 |
0.07 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| TTTCGGGGGCTGTAG............................................................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
15 |
0.07 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................GGAACCTCTGGACTACGCT........................................................................................................................................................................................................................................................................................................................................... |
19 |
3 |
16 |
0.06 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................GTGGAACGGCTTTGTCA............................................................................................................................................................................................................................... |
17 |
2 |
17 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................ACAGGAACCTCTGGT.................................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
17 |
0.06 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................GCAGGAACCTCTGGT.................................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
17 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................GGTGAGCCGGTGGAAAGG............................................................................................................................................................................ |
18 |
2 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................CCGGGAACCTCTGGTCG................................................................................................................................................................................................................................................................................................................................................ |
17 |
2 |
19 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................GGGAACGGCTTTGTCACCG............................................................................................................................................................................................................................ |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................................................................................................................................................GGGGACGGAATCCTGG............ |
16 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................................................CGTTAATAGGTGTGACGGT.......................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................GGAACGGCTTTGTCACCGT........................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................CGGGAACCCCTGGTCGACG............................................................................................................................................................................................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................................................................................................CGGGACCGCCTGAAGGCT......................................................................................... |
18 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| ................................AACCTCTGGTCAACG............................................................................................................................................................................................................................................................................................................................................. |
15 |
1 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................................AGGACTGACAGATACG............................................................................................................................................................................................. |
16 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....GGGGGCTGTAGAAGCCACA..................................................................................................................................................................................................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................................ATAGGGGAGGTTGTTAGCA..................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |