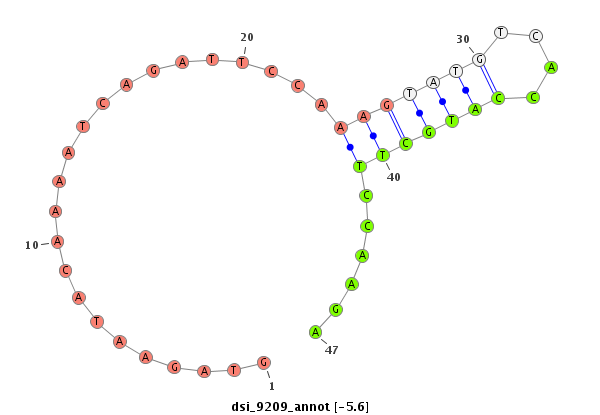
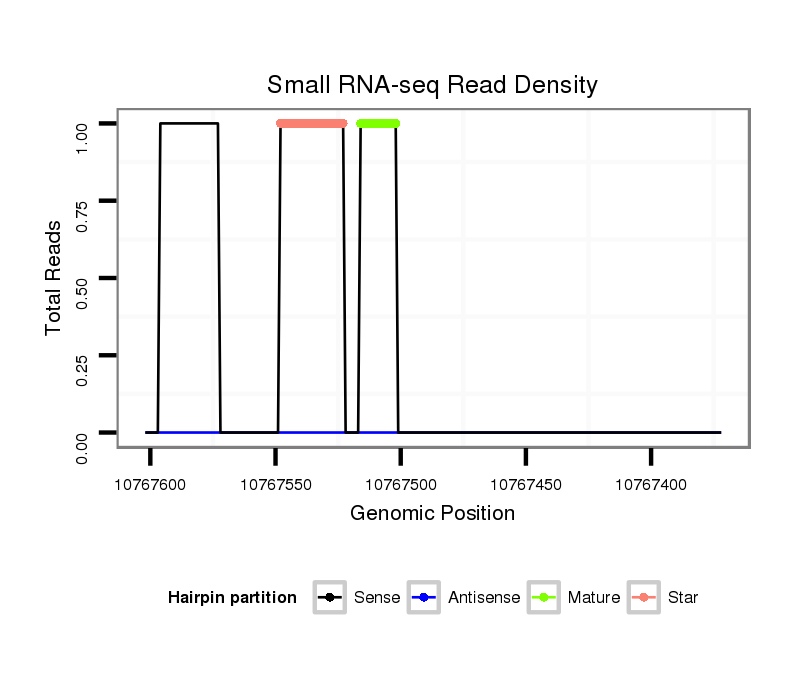
ID:dsi_9209 |
Coordinate:3r:10767422-10767552 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -5.5 | -4.7 | -4.6 |
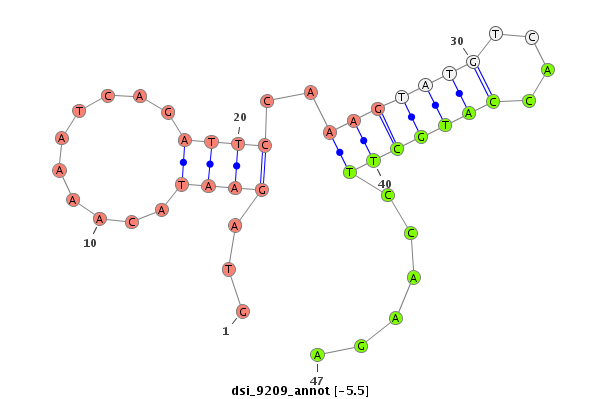 |
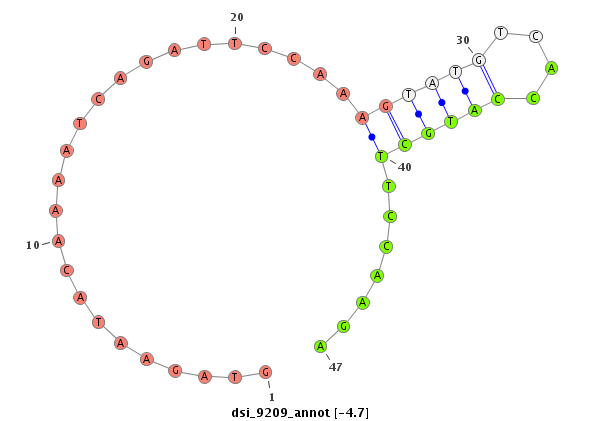 |
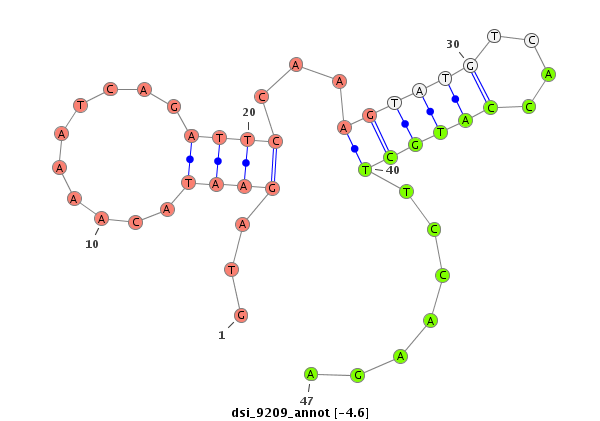 |
exon [3r_10767350_10767421_-]; CDS [3r_10767350_10767421_-]; CDS [3r_10767553_10767610_-]; exon [3r_10767553_10767610_-]; intron [3r_10767422_10767552_-]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| mature | star |
| ##################################################-----------------------------------------------------------------------------------------------------------------------------------################################################## CGCCGTGATGCTAAAGGACTACAAAAAGGCATTGAAATATTGCAAATTGAGTGAGTAGAATACAAAATCAGATTCCAAAGTATGTCACCATGCTTCCAAGAGTTCGCTTTTTTGAGTCTGAAATTCAATCTACTGGGACTTTTATAATGTATAATTTATAATTTTATTCGCCCCGCTGCAGTCCTGCAATACGAGCCCGACAACGCCACGGCCAAAGAGTTCTATCCCCTC ******************************************************.......................(((((((....)))))))......********************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M023 head |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................AAGTAGGTCACCATGCT......................................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ACCATGCTTCCAAGA.................................................................................................................................. | 15 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GATGCTAAAGGACTACAAAAAGGC......................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................GTCGCCATGCTTCGAAGAGTGC.............................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................GTAGAATACAAAATCAGATTCCAAAG....................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........TGCTAAAGGACGATAAAAA............................................................................................................................................................................................................ | 19 | 2 | 5 | 0.60 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................AAAGTAGGTCACCAGGCT......................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TACGGACCCGACAACGCC........................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................TAGAGTTCGTTTTTTTGTGTCTGAA............................................................................................................. | 25 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GCCATGCTTCCAAGA.................................................................................................................................. | 15 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................GATACCATGCTTCCAAGA.................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........TGCTAAACGTTTACAAAAAGG.......................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CCGAAATACGATCCCGACAA............................ | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................CGAAAGTATGTCACC.............................................................................................................................................. | 15 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TATAATGTGGAACTTATAATT.................................................................... | 21 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .GCCGTGATGCGAAAG....................................................................................................................................................................................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................GAAAGTAGGTCACCAGGCT......................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GCGGCACTACGATTTCCTGATGTTTTTCCGTAACTTTATAACGTTTAACTCACTCATCTTATGTTTTAGTCTAAGGTTTCATACAGTGGTACGAAGGTTCTCAAGCGAAAAAACTCAGACTTTAAGTTAGATGACCCTGAAAATATTACATATTAAATATTAAAATAAGCGGGGCGACGTCAGGACGTTATGCTCGGGCTGTTGCGGTGCCGGTTTCTCAAGATAGGGGAG
**********************************************************************************************************************************.......................(((((((....)))))))......****************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................ACGGAGGCTCTCAAGCGA........................................................................................................................... | 18 | 2 | 6 | 0.50 | 3 | 2 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................AGTGGTACGAAGGTG.................................................................................................................................... | 15 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................ATGATACAGTGGTATGAAG....................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................AAGGTACTCAAGCGA........................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................TTAGTCTAAAGTTTTATAGAGT................................................................................................................................................ | 22 | 3 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................TTCACCTAGTGGTACGAAG....................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................AAGGTTTCTTACTGTGG.............................................................................................................................................. | 17 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GTTATGCCCGTGCTGTTGA.......................... | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................GGGCTGTTGCGACGCGGGTT................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:52 AM