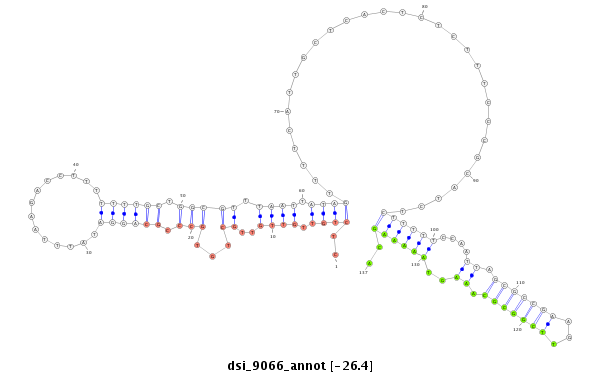




ID:dsi_9066 |
Coordinate:3l:4570451-4570665 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -26.4 | -26.2 | -26.1 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCCCCCGGAGACCAACGCCTCTCCTCCGCAACTTTCTATTACTTTTTGTTAACTTCTCCCCTTCTTTCTTTCGGTTTTCCTCCTGTATTTCTCTCTGTTGTTGTTGCTGTGCCCGCAGGATATTTAAGACCTTTTTTTGCTGGCGTTTAATTATAGTTTTCATTGCTCACTCTCTTTCCCGCATCTCTTTTTCCAATTAGCGCCGAAGTTCGGCGCAAAGTAAAAAGCAAAAATATAGTATTTGTTGTTTTTATTTGGCCCGCAGACCGTGCAGTAAAAACAACAACTTTAACTACAGCAGCAACTTTAACTAAC
********************************************************************************************..((((.((((..((...(((.((((((..............)))))).))))).)))).))))..............................((((((....((.(((((((...))))))).))..))))))..************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
M025 embryo |
SRR618934 dsim w501 ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................................................................................AGTAGAGCAGCAACGTTAA..... | 19 | 3 | 20 | 17.65 | 353 | 353 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TCTTTGTTGTTGTTGCTGTGC........................................................................................................................................................................................................... | 21 | 1 | 1 | 6.00 | 6 | 0 | 0 | 2 | 2 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................CGCAGACCGTACCGTAAA..................................... | 18 | 2 | 2 | 3.50 | 7 | 0 | 2 | 3 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................ACAACAACTTTAACTACAGCGGCA............ | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................AAAACAACAACTTTAACTACAGCGGC............. | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTCGGCGCAAAGTAAAAAGCA...................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................TATGCAGGCGTTTAATTATA................................................................................................................................................................ | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................CTCTGTTGTTGTTGCTGTGCCCGC....................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................CGTGCAGTAAAAACAACAACT........................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TTCTGCCCTTCTCTCGTTCGG................................................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................AGTAGAGCAGCAACTTTAA..... | 19 | 2 | 12 | 0.50 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................CGCAGACCGTACCGTAAAC.................................... | 19 | 3 | 5 | 0.40 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TCGGCGCATAGTAAAA.......................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................GCAGACCGTACCGTAAA..................................... | 17 | 2 | 11 | 0.27 | 3 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................ACGCAGACCGTACCGTAAA..................................... | 19 | 3 | 8 | 0.25 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GTCTTTGTTGTTGTTGCTGTG............................................................................................................................................................................................................ | 21 | 2 | 20 | 0.25 | 5 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................GTTGTTGCGGTGCCCG........................................................................................................................................................................................................ | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................TCTTTGTTGTTGTTGCTGTGCG.......................................................................................................................................................................................................... | 22 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................TGATTGTTAACTCCTCCCCTT............................................................................................................................................................................................................................................................ | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GAGACCAACGCCGCT..................................................................................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................CAACTAGCGCCGAAG........................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................TGTTCACTCGGTTTCCCGCA.................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................CCTTCGGTTTTCCTTCTG...................................................................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................CCCCTCTGTCTTTCGGTT............................................................................................................................................................................................................................................... | 18 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GGAGACTAACGCCGCACC................................................................................................................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................TAAGACCTCTGTTTGC............................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCTGTGGCCGTAGGATA................................................................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................CGTGTAGTAAAAACAAGAG............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GGGGGGCCTCTGGTTGCGGAGAGGAGGCGTTGAAAGATAATGAAAAACAATTGAAGAGGGGAAGAAAGAAAGCCAAAAGGAGGACATAAAGAGAGACAACAACAACGACACGGGCGTCCTATAAATTCTGGAAAAAAACGACCGCAAATTAATATCAAAAGTAACGAGTGAGAGAAAGGGCGTAGAGAAAAAGGTTAATCGCGGCTTCAAGCCGCGTTTCATTTTTCGTTTTTATATCATAAACAACAAAAATAAACCGGGCGTCTGGCACGTCATTTTTGTTGTTGAAATTGATGTCGTCGTTGAAATTGATTG
**************************************************************************************..((((.((((..((...(((.((((((..............)))))).))))).)))).))))..............................((((((....((.(((((((...))))))).))..))))))..******************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR902009 testis |
M024 male body |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................TGAAAGGTAAGGCAAAACAATTG...................................................................................................................................................................................................................................................................... | 23 | 3 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GGGGGGCCTCGGGTT............................................................................................................................................................................................................................................................................................................ | 15 | 1 | 6 | 2.33 | 14 | 0 | 0 | 0 | 9 | 5 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................AACCGGGCGTCTGGCACGTCA........................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................ATGAAAAACAAGTGGAGAGGTGA............................................................................................................................................................................................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................AGAGGAGGCGTTGAAAGACAAT.................................................................................................................................................................................................................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGGTTGCGGAAAGGA.................................................................................................................................................................................................................................................................................................. | 15 | 1 | 5 | 0.80 | 4 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................TGATGTCCTTGGTGAAATTGAT.. | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CCACAACGAAACGGGCGTCCT................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...GGGCCTCGGGTTGCGAAG...................................................................................................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGGCCTCGGGTTG........................................................................................................................................................................................................................................................................................................... | 15 | 1 | 5 | 0.40 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CAGGACATAAAGACAGACAAGA...................................................................................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................TGACGTCGTTGAAATT..... | 16 | 1 | 12 | 0.17 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................CCACAACGAAACGGGCGT...................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| AGGGGGCCTCTGGTT............................................................................................................................................................................................................................................................................................................ | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TGAAAGGTAAGGCAAAACAAT........................................................................................................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GAGAAAAAGGTTAAACTCGGA.............................................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGGCCTCGGGTTTCG......................................................................................................................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGGCCTCGGGTTCAGGA....................................................................................................................................................................................................................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................AAGGAGGACATTAAGCGAA............................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................AAGATAAGGCAAAACAAT........................................................................................................................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGGTTGCGGAAAGTAGCCG.............................................................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AAAAAAACTACCGCTAAT...................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................TGAAAGGTAATGCAAAAC........................................................................................................................................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TATTACCAAAAGTAACGA.................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................................................................................TGGCGTCGTTGAAAT...... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:4570401-4570715 - | dsi_9066 | CCCCCC-GG---AG-ACCAACGCCTCTC------C---------------TCCGCAACTTTCTATTACTTTTT--------------------------------------------GTTA-ACTTCT---------------------CCC---CTTCTTTC-------------------------------------------TTTCGGTTTTCCTCCTGTATTTCT--CTCTGTTGTTGTTGCTGTGCCCGCAGGATATTTAAGAC-----C-TTT-TTTTGCTG--------------------GCGTTTAATTATAGTTTTC----ATT--------------GCTCACTC----------------TCTTTCCCGC------------------A--------TCTCTTTTTCCAATTAGCG-----------CCGAAGTTCGGCGCAAAGTA-AAA---------------------------AGCAAAAATA-----------------TA-----------------------GTATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGCAGTAAAA-ACAACA------ACTTTAAC--------------TACAG-------------------------CAGCAACTTTAACTAAC |
| droSec2 | scaffold_2:4607534-4607827 - | CCCCCC-GG---AG-ACCAACGCCTCTC------------------------CGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCT---------------------CCC---CTTCTTTC-------------------------------------------TTTCGGTTTTCCTCCTGTTTTTCT--CTCTGTTGTTGTTGCTGTGCCCGCAGGATACTTAAGAC--------TT-TTTTGCTG--------------------GCGTTTAATTATAGTTTTG----CTC------------------ACTC----------------TCTTTCCCGC------------------A--------TCTCTTTTTCCAATTAGCG-----------CCGAAGTTCCGCGCAAAGTA-AAA---------------------------AGCAAAAATA-----------------TA-----------------------GTATTTCTTGTTTT--TATTTGGCCCGCAGACCGTGCAGTAAAA-ACAACA------ACTTTAAC--------------TACAG-------------------------CTGCA------------ | |
| dm3 | chr3L:4625300-4625593 - | CCCCTC-GG---AG-ACCAACGCCTCTC------C---------------TTCGCAACTTTCTGTTACTTTAT--------------------------------------------GTTA-ACTTCT---------------------CCC---CTTCTTTC-------------------------------------------TCTTGGTTTTCCTCCTGTATTTTT--CTCTGTTGTTGTTGCTGTGCCCGCAGGATATTTAAGAC-----C-TTT-TTTTGCTG--------------------GCGTTTAATTATAGTTTTC----ATT--------------GCTCTCTC----------------TCCTTCCCGC------------------A--------TCTCTTTTTCCAATTAGCG-----------TCGAAGTTCGGCGTAAAGTA-AAC---------------------------AGCAAAAATA-----------------TA-----------------------GTATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGCAGTAAAA-ACAACA------ACTTTAAC--------------A---------------------------------------------- | |
| droEre2 | scaffold_4784:7335325-7335627 - | CCCTTC-GC---AG-ACCAACGCCTCTC------C---------------TCCGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCT---------------------CCTTAACTTCTTTC-------------------------------------------TCTCGGCCTTCCTTCTTTGTTTCT--CTCTGTTGTTGTTGCTGCGCCCGCAGGATATTTAAGAC-----C-TTT--TTTGCTG--------------------GCGTTTAATTATAGTTTTC----ATT--------------CCCCTCTC----------------TCTTTCCCGC------------------A--------TCTCT--TTCCAATTAGCG-----------CCGAAGTTCAGAGCAAAGTA-AAA---------------------------AGCAAAAATA-----------------TA-----------------------GTATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGCATTAAAT-ACAACA------ACTTTAAC--------------TACAG-------------------------TGGCA------------ | |
| droYak3 | 3L:5200390-5200681 - | CCCCTC-GC---AG-ACCAACGCCTCTC------C---------------TCCGCAGCTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCT--------------------------------GTC-------------------------------------------TCTCGGTTTTCCTACTTTATTTCT--CTCTGTTGTTGTTGCTGTGCCCGCAGGATATTTAAGAC-----C-TTT--TTTGCTG--------------------GCGTTTAATTATAGTTTTC----ATT--------------CCA--CTC----------------TCTTTCCCGC------------------A--------TCTCTTTTTCCAATTAGCG-----------CCGAAGTTCAGCGCAAAGTA-AAA---------------------------AGCAAAAATT-----------------TA-----------------------GTATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGCATTAAAA-ACAACA------ACGTTAAC--------------TACAG-------------------------TGGCA------------ | |
| droEug1 | scf7180000409466:2400637-2400933 - | CCCCTT-GC---AG-ACCAACGCCTCTC------C----------------CTGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCT--------------------TCCC---CTT--------------------------------------------------------TTCCATTTAT-TTTTT--CTTTATTGTTGTTGTTGTGCCCGCAGGATATTTAAGAC-----C-TTT-TTTTGCTG--------------------GCGTTTAATTATAGTTTTT----ATC--------------CTC--CTC----------------TCTTTCTCGC------------------A--------TCTCTTTTTCCAATTAGCG-----------CCGAACTTCGGCGCAAAGTA-A-A---------------------------AGCAAAA-TC-A-AAA--AATC-----TC-----------------------ATATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGAATTAAAA-ACAAC---ACCAACTTTAAC--------------TACAG-------------------------TGGCA-----------G | |
| droBia1 | scf7180000302428:9840528-9840795 + | CGTCTC----------------CCTCCC----------------------CCTGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-GCTGCT---------------------CCC---CTCC--------------------------------------------------CCATTTTCCTCCTT----------TTTGTTGTCGTTCCTGCGCCCGCAGGACCTTCAAGC-----------------CTG--------------------GCGTTTAATTATAGTTTT-----ATC--------------TCC--CTC----------------TCTTTCCCGC------------------A--------ACTC---TTCCAATTAGCG-----------CCGAACTTCAG--CAAAGCA-A-A---------------------------AGCAAAAATC-A---A--AGTCTCATAC------------------------ATATTTGTTGTTT---TATTTGGCCCGCAGACCGTGAATTAAA---CAAC---ATCAACTTTAAC--------------TACAG-------------------------TGGCA-----------G | |
| droTak1 | scf7180000415941:556250-556539 - | CCCCTC-GC---AG-ACCAACGCCTCTC------CCTCTCCTCTCTCTCCACTGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCT---------------------CCC---CTTTTT-------------------------------------------------------------------------------------TTTTGCCCGCAGGATATTTAAGAC-----C-TTTTTTTCGCTG--------------------TCGTTTAATTATAGTTTTT----ATC--------------CCTCTCTC----------------TCTCGCTCGC------------------A--------TCTCT--TTCCAATTAGCG-----------CCGAACTTCGGCACAAAGTA-AAA---------------------------AGCAAAAATC-AAAAA--AATCTC----------------------------ATATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGAATTAAAA-ACAAC---ATCAACTTTAAC--------------TACAG-------------------------TGGTA-----------G | |
| droEle1 | scf7180000491249:5539283-5539593 - | CCCCTT-GC---AG-ACCAACGCCTCTCCTCTCCC---------------TCTGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTTCC---------------------CCC---CTTCTTTT-------------------------------------------TCTTGATTTTCCATA-GT-----------TTTTGTTGTTGTTGTGCCCGCAGGATATTTAAAAC-----C-TTT-TTTCGCTG--------------------GCGTTTAATTATAGTTTTT----CTC--------------CCC--CTC----------------GCCTTCTCGC------------------A--------TCTCTT-TTCCAATTAGCG-----------TCGAACTTTTTGGCAAAGTA-AAA---------------------------AGCAAAAATACTTAAA--AATTTCATAT------------------------ACATATGTTGTTTT-----TTGGCCCGCAGACCCTGAATTAAAA-ACAAC---ATCAAGTTTAAC--------------TACGG-------------------------TGGCA-----------G | |
| droRho1 | scf7180000779636:101142-101436 - | CCCCTT-AC---AG-ACCAACGCCTCTC------CC--------------TCTGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-ACTGCT--------------------CCCC---CTTCATTT-------------------------------------------TCTTGATTTTCCTTC----------------TTTTTGTTGTTGTGCCCGCAGGATATTTAGAAC-----C-TTTTTTTTGCTG--------------------GCGT-TAATTATAGTTTTT----CT-----------------C--CTC----------------TC-TTCTCGC------------------A--------TCTCTT-TTCCAATTAGCG-----------CCGAACTTTAGCCTAAAGTAGAAA---------------------------AGCAAAA-TACAAAAA--AAT-TC----------------------------ATATTTGTTGTTTT----TTTGGCCCGCAGACCGTGAATTAAAA-ACAAC---ATCAAGTTTAAC--------------TACAG-------------------------TGGCA-----------G | |
| droFic1 | scf7180000454065:958701-958991 + | CCC-TT-GC---AG-ACCAACGCCTCTC------C-----------------TGCAACTTTCTGTTACTTTTT--------------------------------------------GTTA-CTTTTT---------------------CCC---CTTCTTTT-----------------------------------------TTTCTTG-TTTTTCTCC-------------------TTTTTGTTGCGCCCGCAGGATATTTAAGAC-----C-CTT-TTTCGCTA--------------------GCGTTTAATTATAGTTTTT----CT----------------------C----------------CCTTTCTCGC------------------A--------TCTCTT-TTCCAATTAGCG-----------CCGAACTTCAGCGCAAAGTAAAAA---------------------------AGCAAAAATACA-AAA--AATCTC----------------------------ATATTTGTTGTTTTTTTATTTGGCCCGCAGACCGTGAATTAAAA-ACAAC---ATCAACTTTACC--------------TACAG-------------------------TGGCA-----------G | |
| droKik1 | scf7180000302486:255290-255563 + | CCCCTC-GC---AGAACCAACGCCTCTC------------------------TGAAACTTTCTGTTACTTT-T--------------------------------------------GTTAAACTTCT--------------------TTCC---CTTCCTC---------------------------------------------------------------TTTCT--CCCTCTTATTGTTGCTGTGCCCGCAGGATATGTGAAAC-----T-TTT-TTCCGCTG--------------------GTGTTAAATTATAGTTTTTTTT-ATC--------------CCT--CTTTCTTTCCTC--------ATCTCCTCC------------------A--------TCCAA-TTTCCAATTAACT-----------ACGAACTTCAGCA-AAAGCA-T-A---------------------------AGCGAAAA-------A--AAT-AT--ATATT---------------------TTATTTGTTGTTT-------------GCAGACCGTGAATTAAAA-ACAACA------------AC--------------TAAAG-------------------------C---------------- | |
| droAna3 | scaffold_13337:2711688-2711999 - | ACACT--------A-ACCAACGCCTCTC------------------------TGCAACTTTCTGTTACTTTTT--------------------------------------------T-------------------------------CCT---CTTCTTTC----------------------------------------------CGTGTTTCCAACTTT-----------TTTTTTTGTTGTTGTGCCCGCAGGATATTAAAGGG-----T-TTT-TTTTTTTG----------------GAACGTGTTTAATTATAGTTTTT----ATTTTTCTTATTTTCTCTCC--CTC----TGTTT----------CGC--TCCATCCCTCCCCTCCCA-C---TATTCAT---ATTTCCCAATTAGCG-----------------------GCAAAGCA-A-A---------------------------GGCAAAAATC-----------------TATT---------------------TCATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGAATAAAAATCCAACA---ACAAC---AAC--------------AAAAT-------------------------CGGCGACTGCGTTTACG | |
| droBip1 | scf7180000396741:28809-29110 + | TTTCGC-AC---TA-ACCAACGCCTCTC-------------------------GCAACTTTCTGTTACTTTTT--------------------------------------------G-------------------------------CCT---CTTCTT-C-------------------------------------------TTCCGTGTTTCCAACTT------------TTTTTTTGTTTTTGTGCCCGCAGGATATTAAAGGC-----T-TTT--TTTTCTG-----------------AACGTGTTTAATTATAGTTTTT----TTC--------------TCT--CTT----TGTTT----------CGC--TCC----------TTCCACC---TATTCAT---ATTTCCCAATTAGCG-----------GCAAACTTTAGGGCAAAGCA-A-A---------------------------GGCAAAAATC-----------------TATT---------------------TCATTTGTTGTTTT--TATTTGGCCCGCAGACCGTGAATAAAAA-CCAACA---GCAAC---AAC--------------AAAAT-------------------------CGGCGACTGCGTTTACG | |
| dp5 | XR_group8:2735148-2735580 - | T-----------------------------------------------------------TCTGTTC-TTGTT--------------------------------------------GTTG-TCG---------------------------------------------------------------TGCTTTCCCCTCCCTTT-CTTCAGTCCTTTTTTTGTGAATTTCC--------TACCCGCTGCGCCCGCAGGATATATCT--CCTGTTCATTT-TTTTGGTGTTCCCTCCCCCCACTCGTACGTGTTTAATTATAGTTTCGCTGCGTC--------------GTTCTCTCTCTTTACTA--------CTCTCCCGC------------------ATTCATATATATCT-CTTCCAATTAGCGGCCCGTGTGCCTCGAATTTCGGTGCAAAGCA-GAACTTCAATAGGGTCTGTGTGTGAG----AGAGAGAGGGAG-AGAAAACT--C---TATTTGATTTAGTTTGATCATAGTTGTATTTGTTGCTTT--TAGTCCCCCTAACGACCGTGAAA-ATAA-GCAACAACATAAAGCTGAAAGCAGAATCAGCATCAACAGAGCGACGCGTCGACGGCGACTGCGACTGCGACTGCGTTTATG | |
| droPer2 | scaffold_24:319212-319780 + | CCCCCGCGCGATAG-ACCAACGCCTCGC------C----------------CAGCAACT-TCTGTTACTTCTTTTGCTCCATCGCGAAGAAATTCTTCGTTTTTTTTTTGTTGTTTTGTGA-ATTTCCTTGTGCCGACGATTCCTTGTCCCA---CTCCATTTATTCATTCTGTTCTTGTTGTTGTCGTGCTTTCCCCTCCCTTT-CTTCAGTCCTTTTTTTGTGAATTTCC--------TACCCGCTGCGCCCGCAGGATATATCT--CCTGTTCA-TT-TTTTGGTGTTCCCTCCCCCCACTCGTACGTGTTTAATTATAGTTTCGCTGCGCC--------------GTTCTCTCGCTTTACTACTCTCCCGCATTCATAC------------------A--------TATCTC-TTCCAATTAGCGGCCCGTGTGCCTCGAATTTCGGTGCAAAGCA-GAACTTCAATACGGTCTGTGTGTGAGAGAGAGAGAGAGGGAG-AGAAAACT--C---TATTTGATTTAGTTTGATCATAGTTGTATTTGTTGCTTT--TAGTCCCCCTAACGACCGTGAAA-ATAA-GCAACAACATAAAGCTGAAAGCAGAATCAGCATCAACAGAGCGACGCGTCGACGGCGAC------TGCGACTGCGTTTAT- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 11:23 AM