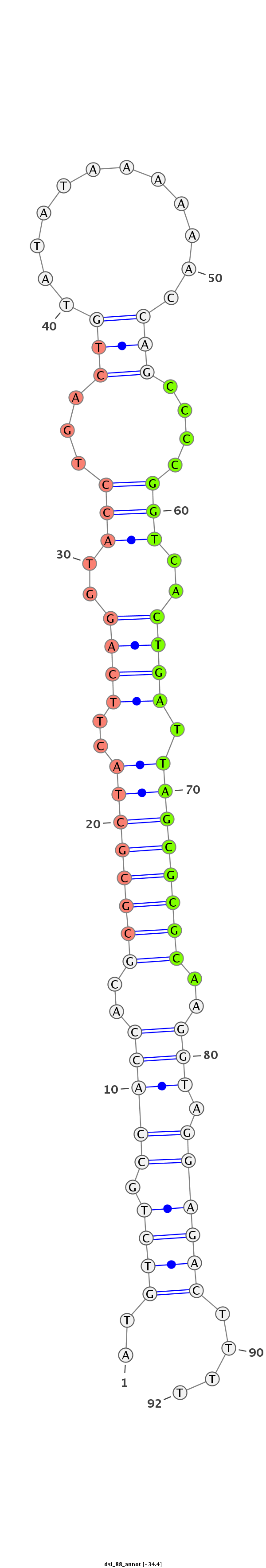
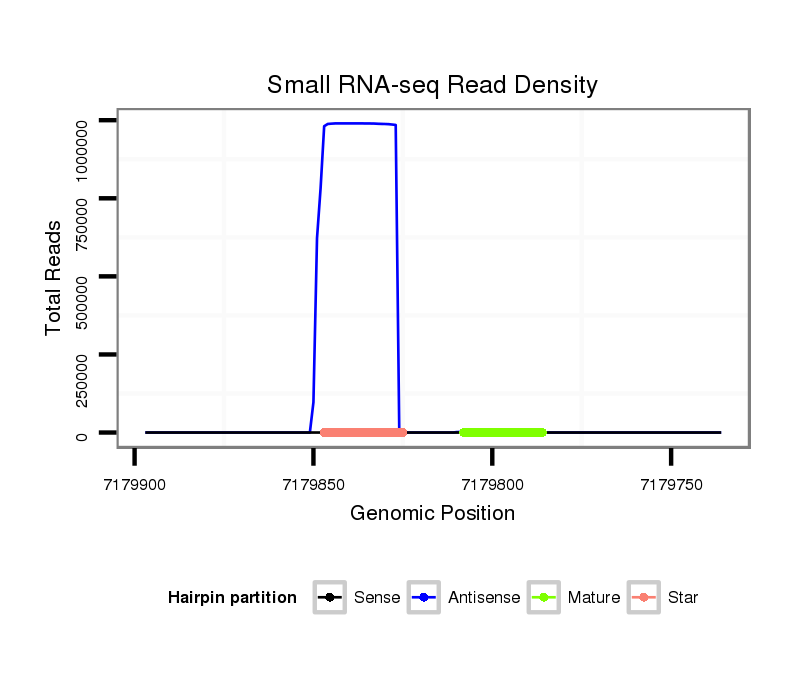
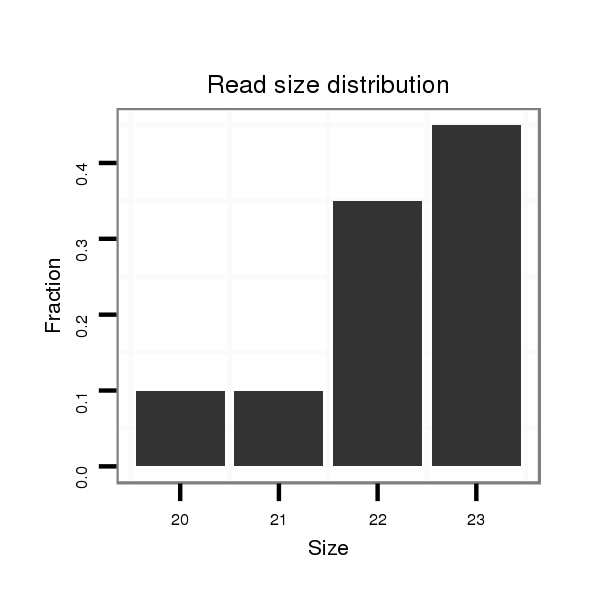
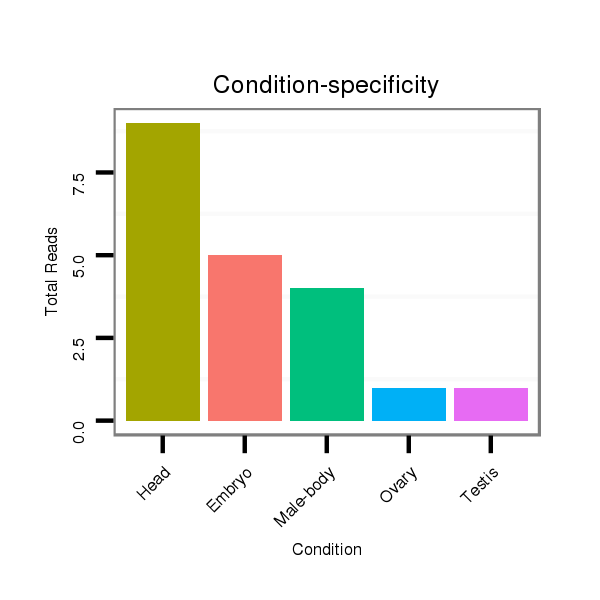

ID:dsi_88 |
Coordinate:2l:7179786-7179847 - |
Confidence:Known Ortholog |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CATTTGGCTTATTTATATGGCGGAGATGGAGATATATGTCTGCCACCACGCGCGCTACTTCAGGTACCTGACTGTATATAAAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTTTCAGCCGGGGGAACATCATCACATATAACGAGTAGG
***********************************..((((.(((((..((((((((..((((..(((...(((............)))....)))..)))).))))))))..))).))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................CCCCGGTCACTGATTAGCGCGCA.................................................. | 23 | 0 | 1 | 8.00 | 8 | 2 | 2 | 3 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................CCCCGGTCACTGATTAGCGCGC................................................... | 22 | 0 | 1 | 7.00 | 7 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CCCCGGTCACTGATTAGCGC..................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ACGCGCGCTACATCAGGTACT.............................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CCCCGGTCACTGATTAGCGCG.................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................CCGGTCACTGATTAGCGCGCA.................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................CCCGGTCACTGATTAGCGCG.................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CGCGCTACTTCAGGTACCTGACT......................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TGACTGTATATAAAAAACCAGCCCCGGTA................................................................. | 29 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TAGACTTTTAAGCCGGG........................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................GTGCCAGCACGCGCGCT.......................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................GCACGCGCGCTACTACTG................................................................................................... | 18 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................GCACGCGCGCTACTTG..................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TCAGGTACCTGACTCTCAA.................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTAAACCGAATAAATATACCGCCTCTACCTCTATATACAGACGGTGGTGCGCGCGATGAAGTCCATGGACTGACATATATTTTTTGGTCGGGGCCAGTGACTAATCGCGCGTTCCATCCTCTGAAAAGTCGGCCCCCTTGTAGTAGTGTATATTGCTCATCC
***********************************..((((.(((((..((((((((..((((..(((...(((............)))....)))..)))).))))))))..))).))))))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
GSM343915 embryo |
M023 head |
M024 male body |
M025 embryo |
SRR1275483 Male prepupae |
O002 Head |
SRR1275485 Male prepupae |
O001 Testis |
SRR1275487 Male larvae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................GCGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 0 | 1 | 523254.00 | 523254 | 116433 | 104175 | 94574 | 115855 | 43983 | 27765 | 16159 | 2251 | 1051 | 196 | 299 | 262 | 91 | 0 | 124 | 0 | 36 |
| ..................................................GCGCGATGAAGTCCATGGACT........................................................................................... | 21 | 0 | 1 | 197740.00 | 197740 | 51276 | 31424 | 22676 | 18195 | 29496 | 24488 | 10410 | 2957 | 4778 | 270 | 414 | 1307 | 40 | 1 | 2 | 0 | 6 |
| .................................................CGCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 0 | 1 | 158428.00 | 158428 | 15739 | 30227 | 22971 | 14218 | 26698 | 20568 | 16046 | 3669 | 2741 | 2236 | 1991 | 1145 | 130 | 0 | 15 | 0 | 34 |
| ...............................................TGCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 0 | 1 | 96723.00 | 96723 | 15799 | 21349 | 20125 | 9988 | 17782 | 10265 | 1115 | 121 | 69 | 18 | 33 | 29 | 22 | 0 | 6 | 0 | 2 |
| ...............................................AGCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 41193.00 | 41193 | 3284 | 9531 | 9437 | 4346 | 8731 | 5243 | 406 | 81 | 64 | 11 | 33 | 20 | 4 | 0 | 0 | 0 | 2 |
| .................................................TGCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 21916.00 | 21916 | 9018 | 3755 | 2609 | 716 | 2886 | 2025 | 526 | 195 | 122 | 21 | 36 | 7 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 16931.00 | 16931 | 1213 | 4140 | 2861 | 405 | 4662 | 3033 | 263 | 141 | 157 | 11 | 22 | 22 | 1 | 0 | 0 | 0 | 0 |
| ...................................................CGCGATGAAGTCCATGGACT........................................................................................... | 20 | 0 | 1 | 7111.00 | 7111 | 360 | 1301 | 960 | 103 | 1068 | 946 | 1004 | 658 | 352 | 125 | 112 | 75 | 37 | 0 | 0 | 0 | 10 |
| ................................................TTGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 2 | 1 | 6572.00 | 6572 | 6302 | 84 | 82 | 54 | 14 | 14 | 11 | 6 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 1 | 1 | 6086.00 | 6086 | 4425 | 465 | 503 | 388 | 183 | 94 | 15 | 1 | 6 | 0 | 4 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................TTTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 2 | 1 | 4599.00 | 4599 | 4328 | 31 | 43 | 157 | 20 | 15 | 2 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................ACGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 3868.00 | 3868 | 528 | 718 | 601 | 816 | 642 | 392 | 116 | 19 | 31 | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................TTTGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 3146.00 | 3146 | 3107 | 5 | 7 | 15 | 5 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................CGCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 2578.00 | 2578 | 129 | 614 | 586 | 416 | 443 | 284 | 83 | 7 | 7 | 2 | 3 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 2 | 1 | 2128.00 | 2128 | 120 | 397 | 339 | 180 | 694 | 373 | 22 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AACGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 2061.00 | 2061 | 44 | 231 | 161 | 521 | 597 | 470 | 19 | 8 | 7 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 1811.00 | 1811 | 879 | 113 | 108 | 562 | 29 | 10 | 84 | 16 | 5 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| .......................................................................................TCGGGGCCAGTGACTAATCGCGC.................................................... | 23 | 0 | 1 | 1526.00 | 1526 | 34 | 290 | 244 | 23 | 131 | 83 | 218 | 56 | 32 | 177 | 154 | 65 | 0 | 7 | 1 | 9 | 2 |
| .............................................AATGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 2 | 1 | 1473.00 | 1473 | 164 | 206 | 154 | 175 | 481 | 286 | 4 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................AAAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 1294.00 | 1294 | 59 | 213 | 171 | 296 | 352 | 185 | 10 | 3 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GCGATGAAGTCCATGGACT........................................................................................... | 19 | 0 | 1 | 1022.00 | 1022 | 235 | 77 | 82 | 14 | 89 | 148 | 163 | 177 | 12 | 11 | 6 | 1 | 6 | 0 | 0 | 0 | 1 |
| ..............................................TAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 2 | 1 | 767.00 | 767 | 298 | 144 | 161 | 45 | 61 | 51 | 3 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTTTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 2 | 1 | 764.00 | 764 | 738 | 1 | 4 | 18 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGTACT........................................................................................... | 23 | 1 | 1 | 681.00 | 681 | 33 | 252 | 222 | 22 | 83 | 39 | 5 | 0 | 0 | 4 | 3 | 15 | 2 | 0 | 0 | 0 | 1 |
| .....................................................CGATGAAGTCCATGGACT........................................................................................... | 18 | 0 | 1 | 677.00 | 677 | 17 | 74 | 79 | 1 | 32 | 27 | 408 | 16 | 0 | 5 | 9 | 0 | 7 | 0 | 0 | 2 | 0 |
| ...............................................TTCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 673.00 | 673 | 558 | 11 | 13 | 67 | 8 | 3 | 3 | 2 | 2 | 1 | 2 | 2 | 0 | 0 | 1 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGAC............................................................................................ | 22 | 0 | 1 | 584.00 | 584 | 157 | 33 | 20 | 158 | 23 | 13 | 12 | 138 | 0 | 1 | 0 | 0 | 23 | 0 | 0 | 0 | 6 |
| ................................................GCGCGCGAGGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 573.00 | 573 | 126 | 100 | 102 | 87 | 85 | 63 | 2 | 4 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GTGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 522.00 | 522 | 92 | 140 | 88 | 69 | 58 | 37 | 34 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TTTCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 2 | 1 | 489.00 | 489 | 476 | 0 | 1 | 11 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGA............................................................................................. | 21 | 0 | 1 | 474.00 | 474 | 66 | 22 | 22 | 48 | 13 | 13 | 274 | 7 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................AAATGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 470.00 | 470 | 13 | 73 | 66 | 67 | 147 | 94 | 7 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGACG........................................................................................... | 23 | 1 | 1 | 470.00 | 470 | 2 | 233 | 223 | 5 | 1 | 1 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................ATGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 1 | 1 | 467.00 | 467 | 316 | 26 | 23 | 63 | 21 | 11 | 4 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCAGGGACT........................................................................................... | 23 | 1 | 1 | 454.00 | 454 | 36 | 115 | 160 | 53 | 41 | 33 | 1 | 3 | 9 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CGGGGCCAGTGACTAATCGCGC.................................................... | 22 | 0 | 1 | 409.00 | 409 | 1 | 9 | 14 | 0 | 1 | 3 | 84 | 9 | 29 | 102 | 72 | 19 | 0 | 30 | 6 | 30 | 0 |
| ................................................GCGCGCGATGAAGTCCATTGACT........................................................................................... | 23 | 1 | 1 | 388.00 | 388 | 34 | 90 | 73 | 54 | 53 | 54 | 26 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGAAGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 384.00 | 384 | 137 | 49 | 46 | 77 | 46 | 23 | 2 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCTATGGACT........................................................................................... | 23 | 1 | 1 | 379.00 | 379 | 71 | 101 | 66 | 78 | 33 | 19 | 9 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGAGT........................................................................................... | 23 | 1 | 1 | 363.00 | 363 | 30 | 107 | 106 | 34 | 7 | 1 | 72 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TTTTGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 3 | 1 | 355.00 | 355 | 353 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCA................................................................................................. | 17 | 0 | 1 | 341.00 | 341 | 0 | 43 | 41 | 0 | 69 | 12 | 176 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGA............................................................................................. | 20 | 0 | 1 | 334.00 | 334 | 5 | 9 | 4 | 3 | 19 | 6 | 242 | 17 | 23 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGACA........................................................................................... | 23 | 1 | 1 | 333.00 | 333 | 11 | 134 | 147 | 16 | 5 | 4 | 13 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GAGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 330.00 | 330 | 92 | 68 | 41 | 76 | 16 | 19 | 16 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCAT................................................................................................ | 18 | 0 | 1 | 326.00 | 326 | 0 | 83 | 81 | 0 | 108 | 20 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................ATGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 2 | 1 | 323.00 | 323 | 285 | 11 | 9 | 6 | 3 | 3 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGAC............................................................................................ | 21 | 0 | 1 | 319.00 | 319 | 18 | 13 | 6 | 20 | 8 | 9 | 24 | 202 | 4 | 2 | 6 | 0 | 4 | 0 | 0 | 1 | 2 |
| .................................................CGCGCGAGGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 304.00 | 304 | 19 | 39 | 38 | 12 | 108 | 65 | 2 | 1 | 10 | 2 | 5 | 3 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATTAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 297.00 | 297 | 36 | 63 | 61 | 17 | 68 | 43 | 5 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGTACT........................................................................................... | 22 | 1 | 1 | 294.00 | 294 | 3 | 67 | 65 | 3 | 45 | 29 | 9 | 3 | 0 | 30 | 15 | 25 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCTATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 294.00 | 294 | 25 | 80 | 76 | 28 | 39 | 31 | 9 | 1 | 1 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGTACT........................................................................................... | 21 | 1 | 1 | 290.00 | 290 | 9 | 61 | 55 | 8 | 52 | 19 | 0 | 0 | 2 | 3 | 7 | 74 | 0 | 0 | 0 | 0 | 0 |
| ................................................CCGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 282.00 | 282 | 26 | 59 | 50 | 52 | 30 | 20 | 33 | 0 | 5 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGTGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 282.00 | 282 | 25 | 85 | 64 | 27 | 42 | 21 | 14 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGA............................................................................................. | 19 | 0 | 1 | 279.00 | 279 | 13 | 5 | 1 | 4 | 5 | 6 | 198 | 10 | 34 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................ACGCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 270.00 | 270 | 43 | 45 | 35 | 13 | 43 | 45 | 28 | 10 | 5 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCAAGGACT........................................................................................... | 23 | 1 | 1 | 265.00 | 265 | 34 | 61 | 84 | 63 | 9 | 6 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGAGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 261.00 | 261 | 75 | 41 | 40 | 45 | 21 | 9 | 17 | 8 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................GCGCGCGATGAAGTCCATGGGCT........................................................................................... | 23 | 1 | 1 | 251.00 | 251 | 50 | 62 | 45 | 28 | 18 | 14 | 29 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGTGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 249.00 | 249 | 51 | 45 | 28 | 65 | 14 | 13 | 29 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TATGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 2 | 1 | 245.00 | 245 | 211 | 3 | 2 | 28 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GGCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 244.00 | 244 | 22 | 56 | 60 | 15 | 51 | 13 | 6 | 3 | 0 | 6 | 3 | 9 | 0 | 0 | 0 | 0 | 0 |
| ................................................AAGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 2 | 1 | 242.00 | 242 | 32 | 49 | 28 | 4 | 78 | 41 | 6 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................GGGGCCAGTGACTAATCGCGC.................................................... | 21 | 0 | 1 | 240.00 | 240 | 0 | 55 | 46 | 0 | 16 | 13 | 51 | 13 | 4 | 8 | 5 | 2 | 0 | 7 | 10 | 10 | 0 |
| ..................................................GCGCGATGAAGTCCATGGAC............................................................................................ | 20 | 0 | 1 | 239.00 | 239 | 39 | 1 | 2 | 10 | 5 | 4 | 7 | 163 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................ATTGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 3 | 1 | 221.00 | 221 | 213 | 1 | 1 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCAGGGACT........................................................................................... | 21 | 1 | 1 | 218.00 | 218 | 35 | 38 | 37 | 7 | 15 | 12 | 1 | 7 | 50 | 3 | 3 | 10 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGAGGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 211.00 | 211 | 11 | 32 | 30 | 11 | 57 | 47 | 1 | 4 | 14 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................TTTTCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 208.00 | 208 | 207 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCTCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 208.00 | 208 | 29 | 44 | 52 | 30 | 10 | 16 | 4 | 15 | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGG.............................................................................................. | 19 | 0 | 1 | 208.00 | 208 | 0 | 1 | 0 | 0 | 3 | 0 | 204 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTTAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 207.00 | 207 | 200 | 1 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................AAACGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 3 | 1 | 205.00 | 205 | 5 | 21 | 15 | 55 | 66 | 38 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGACC........................................................................................... | 23 | 1 | 1 | 203.00 | 203 | 14 | 64 | 63 | 13 | 11 | 7 | 26 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GGGCCAGTGACTAATCGCGC.................................................... | 20 | 0 | 1 | 203.00 | 203 | 2 | 49 | 18 | 0 | 9 | 7 | 16 | 5 | 1 | 4 | 1 | 0 | 0 | 53 | 0 | 38 | 0 |
| ................................................GCGCTCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 201.00 | 201 | 28 | 47 | 59 | 18 | 20 | 16 | 9 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................ATTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 2 | 1 | 199.00 | 199 | 145 | 5 | 3 | 36 | 3 | 2 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGCCCATGGACT........................................................................................... | 23 | 1 | 1 | 196.00 | 196 | 18 | 40 | 34 | 27 | 34 | 12 | 21 | 3 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCAGGGACT........................................................................................... | 22 | 1 | 1 | 192.00 | 192 | 10 | 33 | 44 | 15 | 22 | 22 | 1 | 6 | 22 | 5 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGAAT........................................................................................... | 23 | 1 | 1 | 191.00 | 191 | 40 | 20 | 16 | 50 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TATTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 2 | 1 | 190.00 | 190 | 184 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCACGGACT........................................................................................... | 23 | 1 | 1 | 186.00 | 186 | 32 | 35 | 31 | 39 | 22 | 8 | 15 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................GCACGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 181.00 | 181 | 31 | 42 | 41 | 29 | 7 | 7 | 22 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GGGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 180.00 | 180 | 51 | 29 | 11 | 52 | 10 | 13 | 10 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGACGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 179.00 | 179 | 44 | 36 | 28 | 26 | 15 | 10 | 14 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................TTAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 176.00 | 176 | 154 | 4 | 7 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGACT............................................................................................ | 22 | 3 | 1 | 170.00 | 170 | 32 | 24 | 17 | 25 | 38 | 19 | 9 | 1 | 0 | 2 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................AATTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 166.00 | 166 | 23 | 16 | 19 | 35 | 47 | 24 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCA................................................................................................. | 16 | 0 | 1 | 165.00 | 165 | 0 | 1 | 1 | 0 | 6 | 3 | 154 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGATT........................................................................................... | 23 | 1 | 1 | 165.00 | 165 | 32 | 17 | 20 | 15 | 7 | 11 | 60 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCCATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 163.00 | 163 | 13 | 46 | 32 | 15 | 33 | 15 | 6 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGGTGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 163.00 | 163 | 20 | 45 | 47 | 20 | 6 | 4 | 10 | 3 | 2 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGAACT........................................................................................... | 23 | 1 | 1 | 162.00 | 162 | 40 | 21 | 14 | 56 | 12 | 6 | 12 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGAAGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 162.00 | 162 | 19 | 13 | 15 | 12 | 65 | 30 | 4 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGACG........................................................................................... | 21 | 1 | 1 | 161.00 | 161 | 1 | 85 | 63 | 2 | 1 | 2 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGAGT........................................................................................... | 22 | 1 | 1 | 159.00 | 159 | 4 | 52 | 26 | 5 | 4 | 3 | 63 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGTAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 159.00 | 159 | 125 | 3 | 1 | 23 | 3 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGATT........................................................................................... | 22 | 1 | 1 | 157.00 | 157 | 15 | 20 | 25 | 2 | 25 | 10 | 44 | 10 | 1 | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCACGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 157.00 | 157 | 49 | 12 | 14 | 35 | 19 | 8 | 19 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................TAGCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 2 | 1 | 151.00 | 151 | 106 | 13 | 11 | 5 | 7 | 4 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGG.............................................................................................. | 18 | 0 | 1 | 151.00 | 151 | 0 | 0 | 0 | 0 | 0 | 0 | 148 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGG.............................................................................................. | 20 | 0 | 1 | 150.00 | 150 | 0 | 0 | 0 | 0 | 0 | 0 | 150 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATTGACT........................................................................................... | 22 | 1 | 1 | 149.00 | 149 | 1 | 20 | 13 | 8 | 32 | 20 | 22 | 0 | 0 | 9 | 8 | 16 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCTCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 148.00 | 148 | 7 | 38 | 31 | 6 | 24 | 23 | 4 | 7 | 3 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCTATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 146.00 | 146 | 3 | 24 | 22 | 1 | 47 | 31 | 9 | 0 | 4 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCGCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 138.00 | 138 | 60 | 11 | 9 | 13 | 7 | 1 | 3 | 32 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AAAGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 3 | 1 | 138.00 | 138 | 5 | 16 | 11 | 5 | 55 | 40 | 1 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................GCCAGTGACTAATCGCGC.................................................... | 18 | 0 | 1 | 137.00 | 137 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 1 | 0 | 25 | 10 | 1 | 0 | 48 | 1 | 46 | 1 |
| .................................................CGCGCGATGAAGTCCATG............................................................................................... | 18 | 0 | 1 | 136.00 | 136 | 0 | 13 | 6 | 0 | 19 | 1 | 94 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ATCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 135.00 | 135 | 29 | 4 | 1 | 68 | 2 | 1 | 23 | 1 | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TTTTTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 28 | 3 | 1 | 135.00 | 135 | 133 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAATCCATGGACT........................................................................................... | 23 | 1 | 1 | 134.00 | 134 | 19 | 40 | 33 | 19 | 4 | 12 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCTATGGACT........................................................................................... | 22 | 1 | 1 | 133.00 | 133 | 10 | 36 | 21 | 3 | 24 | 18 | 16 | 1 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATTGACT........................................................................................... | 21 | 1 | 1 | 132.00 | 132 | 15 | 21 | 17 | 5 | 28 | 22 | 15 | 2 | 0 | 0 | 1 | 6 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTTCATGGACT........................................................................................... | 23 | 1 | 1 | 131.00 | 131 | 14 | 40 | 21 | 8 | 18 | 19 | 10 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AATGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 3 | 1 | 131.00 | 131 | 63 | 14 | 7 | 4 | 20 | 20 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCTATGGACT........................................................................................... | 21 | 1 | 1 | 127.00 | 127 | 17 | 33 | 22 | 18 | 12 | 17 | 3 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATTAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 126.00 | 126 | 11 | 17 | 7 | 5 | 45 | 32 | 6 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 123.00 | 123 | 18 | 12 | 7 | 14 | 9 | 2 | 5 | 5 | 5 | 11 | 24 | 11 | 0 | 0 | 0 | 0 | 0 |
| .............................................TAAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 122.00 | 122 | 86 | 8 | 10 | 10 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCCCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 122.00 | 122 | 22 | 8 | 10 | 10 | 48 | 17 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| .................................................CCCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 122.00 | 122 | 4 | 13 | 12 | 8 | 53 | 17 | 9 | 4 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGTGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 121.00 | 121 | 10 | 41 | 22 | 8 | 22 | 16 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGAGT........................................................................................... | 21 | 1 | 1 | 120.00 | 120 | 8 | 27 | 31 | 7 | 1 | 2 | 40 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGAGGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 120.00 | 120 | 28 | 30 | 20 | 6 | 24 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 120.00 | 120 | 23 | 17 | 13 | 3 | 23 | 19 | 10 | 3 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGACG........................................................................................... | 22 | 1 | 1 | 119.00 | 119 | 0 | 54 | 49 | 1 | 1 | 0 | 10 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................GCGCGAAGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 117.00 | 117 | 28 | 14 | 11 | 10 | 20 | 24 | 2 | 2 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CCGCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 116.00 | 116 | 10 | 12 | 12 | 14 | 31 | 11 | 18 | 1 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGCACT........................................................................................... | 23 | 1 | 1 | 114.00 | 114 | 9 | 11 | 12 | 14 | 38 | 19 | 5 | 0 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGACA........................................................................................... | 22 | 1 | 1 | 114.00 | 114 | 2 | 51 | 41 | 1 | 3 | 2 | 8 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGGCT........................................................................................... | 22 | 1 | 1 | 113.00 | 113 | 6 | 19 | 9 | 9 | 18 | 13 | 25 | 5 | 4 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAATTCCATGGACT........................................................................................... | 23 | 1 | 1 | 112.00 | 112 | 17 | 33 | 28 | 15 | 10 | 6 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGTACT........................................................................................... | 24 | 1 | 1 | 111.00 | 111 | 0 | 38 | 29 | 3 | 33 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCCCGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 111.00 | 111 | 14 | 29 | 23 | 14 | 19 | 7 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGCCAGTGACTAATCGCGC.................................................... | 19 | 0 | 1 | 110.00 | 110 | 0 | 5 | 3 | 0 | 2 | 4 | 2 | 6 | 0 | 6 | 1 | 0 | 0 | 44 | 1 | 36 | 0 |
| ...............................................TACGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 110.00 | 110 | 41 | 17 | 16 | 11 | 8 | 14 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGCTGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 109.00 | 109 | 1 | 18 | 29 | 4 | 31 | 18 | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGGCCATGGACT........................................................................................... | 23 | 1 | 1 | 108.00 | 108 | 4 | 53 | 39 | 3 | 5 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CTCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 106.00 | 106 | 12 | 14 | 6 | 6 | 8 | 18 | 8 | 23 | 2 | 7 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAGGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 106.00 | 106 | 12 | 23 | 26 | 10 | 5 | 6 | 19 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGGCT........................................................................................... | 21 | 1 | 1 | 106.00 | 106 | 17 | 8 | 14 | 6 | 19 | 13 | 12 | 5 | 11 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATG............................................................................................... | 19 | 0 | 1 | 103.00 | 103 | 0 | 7 | 1 | 0 | 6 | 5 | 84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGAAT........................................................................................... | 22 | 1 | 1 | 103.00 | 103 | 3 | 6 | 5 | 4 | 2 | 2 | 75 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGACA........................................................................................... | 21 | 1 | 1 | 102.00 | 102 | 8 | 47 | 29 | 0 | 3 | 5 | 3 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGACG........................................................................................... | 24 | 1 | 1 | 102.00 | 102 | 1 | 51 | 48 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCAGGGACT........................................................................................... | 24 | 1 | 1 | 99.00 | 99 | 10 | 28 | 28 | 4 | 14 | 14 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGACT............................................................................................ | 21 | 3 | 1 | 99.00 | 99 | 5 | 7 | 5 | 4 | 26 | 9 | 20 | 0 | 0 | 9 | 4 | 10 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TTCGGGGCCAGTGACTAATCGCGC.................................................... | 24 | 1 | 1 | 99.00 | 99 | 14 | 6 | 7 | 4 | 4 | 2 | 6 | 11 | 4 | 18 | 20 | 1 | 0 | 0 | 1 | 0 | 1 |
| ................................................GCGCGCGATGAAGTCCATCGACT........................................................................................... | 23 | 1 | 1 | 98.00 | 98 | 14 | 17 | 13 | 10 | 23 | 11 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCC.................................................................................................. | 16 | 0 | 1 | 98.00 | 98 | 0 | 15 | 7 | 0 | 10 | 14 | 52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCACGGACT........................................................................................... | 22 | 1 | 1 | 98.00 | 98 | 8 | 8 | 6 | 5 | 11 | 9 | 18 | 9 | 19 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGAGGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 98.00 | 98 | 3 | 23 | 13 | 2 | 40 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGGAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 97.00 | 97 | 15 | 16 | 6 | 14 | 8 | 4 | 27 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGCGATGAAGTCCATGGACT........................................................................................... | 20 | 1 | 1 | 97.00 | 97 | 84 | 1 | 2 | 1 | 2 | 2 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCAAGGACT........................................................................................... | 21 | 1 | 1 | 97.00 | 97 | 50 | 14 | 11 | 6 | 3 | 4 | 1 | 5 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CACGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 97.00 | 97 | 4 | 13 | 4 | 3 | 11 | 9 | 23 | 1 | 0 | 12 | 14 | 3 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGAGCGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 96.00 | 96 | 26 | 14 | 9 | 31 | 1 | 7 | 6 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATCAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 94.00 | 94 | 5 | 18 | 17 | 7 | 30 | 11 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GAGCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 93.00 | 93 | 22 | 15 | 6 | 5 | 11 | 17 | 8 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCA................................................................................................. | 15 | 0 | 1 | 92.00 | 92 | 0 | 5 | 5 | 0 | 9 | 4 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGACC........................................................................................... | 22 | 1 | 1 | 91.00 | 91 | 1 | 21 | 16 | 0 | 7 | 8 | 26 | 5 | 3 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGAAGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 91.00 | 91 | 27 | 14 | 11 | 13 | 17 | 7 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCACGGACT........................................................................................... | 21 | 1 | 1 | 90.00 | 90 | 18 | 10 | 5 | 3 | 12 | 7 | 11 | 5 | 18 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATAAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 90.00 | 90 | 22 | 9 | 6 | 18 | 15 | 9 | 10 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGACCATGGACT........................................................................................... | 23 | 1 | 1 | 90.00 | 90 | 6 | 46 | 25 | 5 | 2 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGCACT........................................................................................... | 22 | 1 | 1 | 89.00 | 89 | 2 | 7 | 4 | 0 | 29 | 13 | 6 | 2 | 1 | 12 | 4 | 8 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGACT............................................................................................ | 20 | 3 | 1 | 89.00 | 89 | 14 | 10 | 5 | 1 | 12 | 17 | 4 | 1 | 0 | 1 | 1 | 23 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGGGATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 88.00 | 88 | 3 | 28 | 22 | 4 | 14 | 12 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGCCCATGGACT........................................................................................... | 22 | 1 | 1 | 88.00 | 88 | 1 | 15 | 5 | 2 | 13 | 13 | 32 | 1 | 1 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGATT........................................................................................... | 21 | 1 | 1 | 86.00 | 86 | 5 | 5 | 7 | 1 | 9 | 11 | 41 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAATTCCATGGACT........................................................................................... | 21 | 1 | 1 | 85.00 | 85 | 8 | 20 | 9 | 6 | 21 | 13 | 5 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCCATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 85.00 | 85 | 1 | 12 | 9 | 1 | 37 | 17 | 2 | 1 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGAGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 85.00 | 85 | 13 | 9 | 13 | 2 | 13 | 12 | 6 | 8 | 7 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| ................................................GCGCGCAATGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 84.00 | 84 | 16 | 10 | 11 | 16 | 8 | 11 | 11 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGGTGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 83.00 | 83 | 0 | 19 | 17 | 3 | 5 | 5 | 15 | 7 | 3 | 4 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGACC........................................................................................... | 21 | 1 | 1 | 83.00 | 83 | 7 | 20 | 12 | 4 | 8 | 13 | 10 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCTATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 83.00 | 83 | 3 | 21 | 20 | 4 | 17 | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTTCATGGACT........................................................................................... | 22 | 1 | 1 | 82.00 | 82 | 0 | 21 | 12 | 2 | 22 | 15 | 7 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGAGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 81.00 | 81 | 26 | 10 | 10 | 2 | 7 | 4 | 2 | 13 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGAAT........................................................................................... | 21 | 1 | 1 | 81.00 | 81 | 20 | 8 | 3 | 7 | 1 | 1 | 30 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCC.................................................................................................. | 15 | 0 | 1 | 80.00 | 80 | 0 | 3 | 2 | 0 | 5 | 15 | 55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGTGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 80.00 | 80 | 3 | 18 | 10 | 7 | 7 | 11 | 20 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AACGCGATGAAGTCCATGGACT........................................................................................... | 22 | 2 | 1 | 78.00 | 78 | 4 | 6 | 7 | 3 | 35 | 21 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGAGT........................................................................................... | 24 | 1 | 1 | 78.00 | 78 | 3 | 33 | 34 | 4 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATTAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 78.00 | 78 | 5 | 11 | 8 | 1 | 24 | 15 | 6 | 2 | 0 | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCTATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 77.00 | 77 | 8 | 11 | 10 | 6 | 13 | 14 | 7 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................GCGCGATGAAGCCCATGGACT........................................................................................... | 21 | 1 | 1 | 76.00 | 76 | 10 | 10 | 5 | 3 | 17 | 13 | 12 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTTCATGGACT........................................................................................... | 21 | 1 | 1 | 75.00 | 75 | 5 | 15 | 6 | 5 | 19 | 20 | 2 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................CAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 2 | 1 | 74.00 | 74 | 8 | 9 | 25 | 19 | 6 | 3 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGACGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 74.00 | 74 | 4 | 2 | 2 | 1 | 0 | 3 | 1 | 0 | 61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATAGACT........................................................................................... | 23 | 1 | 1 | 73.00 | 73 | 8 | 11 | 11 | 14 | 13 | 4 | 9 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TATGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 72.00 | 72 | 69 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAACTCCATGGACT........................................................................................... | 23 | 1 | 1 | 72.00 | 72 | 10 | 19 | 18 | 12 | 10 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGAC............................................................................................ | 23 | 0 | 1 | 71.00 | 71 | 20 | 2 | 3 | 12 | 5 | 2 | 19 | 5 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATG............................................................................................... | 17 | 0 | 1 | 70.00 | 70 | 0 | 1 | 1 | 0 | 3 | 1 | 64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGACA........................................................................................... | 24 | 1 | 1 | 70.00 | 70 | 3 | 41 | 24 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGGTGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 69.00 | 69 | 5 | 18 | 13 | 1 | 5 | 6 | 11 | 1 | 7 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................GCCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 2 | 1 | 69.00 | 69 | 18 | 10 | 6 | 20 | 7 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGACGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 69.00 | 69 | 3 | 13 | 5 | 4 | 9 | 7 | 15 | 4 | 7 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAGGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 68.00 | 68 | 2 | 7 | 9 | 1 | 9 | 4 | 16 | 0 | 19 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGTGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 68.00 | 68 | 3 | 17 | 28 | 2 | 11 | 6 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCAAGGACT........................................................................................... | 22 | 1 | 1 | 68.00 | 68 | 7 | 15 | 16 | 10 | 3 | 4 | 1 | 5 | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................CGCGTGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 67.00 | 67 | 4 | 13 | 5 | 6 | 11 | 8 | 12 | 3 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAGGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 67.00 | 67 | 0 | 8 | 9 | 0 | 10 | 6 | 26 | 0 | 5 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCTATGGACT........................................................................................... | 24 | 1 | 1 | 67.00 | 67 | 14 | 17 | 10 | 9 | 9 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTACATGGACT........................................................................................... | 23 | 1 | 1 | 66.00 | 66 | 14 | 7 | 8 | 23 | 2 | 0 | 1 | 6 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGACGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 66.00 | 66 | 5 | 2 | 6 | 6 | 15 | 7 | 13 | 3 | 5 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCA................................................................................................. | 18 | 0 | 1 | 65.00 | 65 | 0 | 8 | 5 | 0 | 33 | 6 | 12 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCCCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 65.00 | 65 | 2 | 6 | 3 | 2 | 32 | 13 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................AGCGATGAAGTCCATGGACT........................................................................................... | 20 | 1 | 1 | 65.00 | 65 | 11 | 15 | 11 | 0 | 11 | 14 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGCACT........................................................................................... | 21 | 1 | 1 | 64.00 | 64 | 3 | 4 | 5 | 0 | 17 | 4 | 2 | 1 | 1 | 0 | 1 | 26 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGTGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 64.00 | 64 | 9 | 10 | 6 | 5 | 7 | 10 | 7 | 1 | 8 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCAT................................................................................................ | 19 | 1 | 1 | 62.00 | 62 | 0 | 12 | 13 | 0 | 34 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................CCGGGGCCAGTGACTAATCGCGC.................................................... | 23 | 1 | 1 | 61.00 | 61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 20 | 13 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGA............................................................................................. | 22 | 0 | 1 | 61.00 | 61 | 10 | 5 | 5 | 4 | 4 | 8 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGCGCGAGGAAGTCCATGGACT........................................................................................... | 22 | 2 | 1 | 61.00 | 61 | 10 | 12 | 7 | 0 | 23 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATTGACT........................................................................................... | 24 | 1 | 1 | 60.00 | 60 | 2 | 16 | 10 | 1 | 16 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCACGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 59.00 | 59 | 11 | 3 | 2 | 3 | 11 | 5 | 18 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGACTC.......................................................................................... | 24 | 1 | 1 | 59.00 | 59 | 18 | 1 | 2 | 24 | 4 | 0 | 8 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCATGTACT........................................................................................... | 24 | 2 | 1 | 59.00 | 59 | 2 | 23 | 16 | 0 | 15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCAATGGACT........................................................................................... | 23 | 1 | 1 | 59.00 | 59 | 12 | 12 | 8 | 15 | 3 | 2 | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGGACTC.......................................................................................... | 22 | 1 | 1 | 58.00 | 58 | 8 | 1 | 1 | 4 | 0 | 2 | 32 | 3 | 6 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCCCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 58.00 | 58 | 1 | 0 | 4 | 0 | 32 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCAT................................................................................................ | 17 | 0 | 1 | 58.00 | 58 | 0 | 0 | 2 | 0 | 6 | 4 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................ATAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 58.00 | 58 | 36 | 2 | 0 | 13 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCAT................................................................................................ | 16 | 0 | 1 | 57.00 | 57 | 0 | 7 | 10 | 0 | 15 | 6 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................TCGCGCGATGAAGTCCATGGAC............................................................................................ | 22 | 1 | 1 | 57.00 | 57 | 17 | 4 | 2 | 14 | 3 | 3 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCAATGGACT........................................................................................... | 21 | 1 | 1 | 57.00 | 57 | 6 | 10 | 9 | 2 | 5 | 5 | 0 | 3 | 14 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCGATGGACT........................................................................................... | 23 | 1 | 1 | 57.00 | 57 | 8 | 21 | 15 | 5 | 5 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCGTGGACT........................................................................................... | 23 | 1 | 1 | 55.00 | 55 | 12 | 6 | 2 | 6 | 10 | 2 | 15 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCCATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 55.00 | 55 | 1 | 11 | 10 | 1 | 24 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCACGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 55.00 | 55 | 6 | 5 | 4 | 2 | 7 | 1 | 16 | 2 | 4 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................GCGCGCGATGAAGTGCATGGACT........................................................................................... | 23 | 1 | 1 | 54.00 | 54 | 7 | 20 | 9 | 9 | 2 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGGAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 54.00 | 54 | 0 | 2 | 3 | 1 | 6 | 8 | 25 | 3 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTC................................................................................................... | 15 | 0 | 1 | 53.00 | 53 | 0 | 8 | 3 | 0 | 6 | 13 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCTCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 53.00 | 53 | 6 | 8 | 18 | 0 | 11 | 5 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................ATTTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 53.00 | 53 | 50 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGT.................................................................................................... | 15 | 0 | 1 | 52.00 | 52 | 0 | 5 | 15 | 0 | 24 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATCGACT........................................................................................... | 21 | 1 | 1 | 52.00 | 52 | 4 | 3 | 6 | 0 | 18 | 7 | 8 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................TTCGCGATGAAGTCCATGGACT........................................................................................... | 22 | 2 | 2 | 51.50 | 103 | 89 | 5 | 2 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCATTGACT........................................................................................... | 24 | 2 | 1 | 51.00 | 51 | 4 | 10 | 8 | 2 | 20 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGCTGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 51.00 | 51 | 11 | 5 | 3 | 5 | 7 | 1 | 2 | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATTAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 50.00 | 50 | 2 | 13 | 10 | 3 | 17 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGAAGTCCATGGTCT........................................................................................... | 23 | 1 | 1 | 50.00 | 50 | 10 | 5 | 18 | 7 | 4 | 1 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTCCATGAACT........................................................................................... | 21 | 1 | 1 | 50.00 | 50 | 21 | 2 | 3 | 10 | 6 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CGCGATGAAGTCCATGG.............................................................................................. | 17 | 0 | 1 | 50.00 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGACGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 50.00 | 50 | 4 | 15 | 6 | 3 | 5 | 1 | 2 | 0 | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCA................................................................................................. | 18 | 1 | 1 | 49.00 | 49 | 0 | 7 | 10 | 0 | 21 | 4 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCATGGACG........................................................................................... | 24 | 2 | 1 | 49.00 | 49 | 0 | 26 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGTGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 48.00 | 48 | 3 | 12 | 13 | 5 | 7 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCTCGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 48.00 | 48 | 7 | 2 | 7 | 4 | 5 | 9 | 4 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCAATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 47.00 | 47 | 2 | 5 | 1 | 1 | 5 | 7 | 16 | 4 | 4 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCAT................................................................................................ | 19 | 0 | 1 | 47.00 | 47 | 0 | 8 | 10 | 0 | 23 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCTCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 47.00 | 47 | 2 | 12 | 5 | 0 | 18 | 8 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TATAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 46.00 | 46 | 39 | 0 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ATCGGGGCCAGTGACTAATCGCGC.................................................... | 24 | 1 | 1 | 46.00 | 46 | 6 | 11 | 2 | 6 | 9 | 2 | 8 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAGTACATGGACT........................................................................................... | 21 | 1 | 1 | 46.00 | 46 | 17 | 4 | 5 | 5 | 1 | 0 | 0 | 4 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAATCCATGGACT........................................................................................... | 22 | 1 | 1 | 45.00 | 45 | 3 | 8 | 8 | 10 | 2 | 1 | 4 | 5 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGACC........................................................................................... | 24 | 1 | 1 | 45.00 | 45 | 1 | 14 | 16 | 3 | 5 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCCATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 44.00 | 44 | 4 | 10 | 4 | 1 | 15 | 5 | 1 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGGAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 44.00 | 44 | 6 | 3 | 4 | 4 | 1 | 4 | 18 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGAGCGATGAAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 44.00 | 44 | 4 | 14 | 7 | 1 | 6 | 6 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................TGCGTGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 43.00 | 43 | 4 | 7 | 4 | 3 | 16 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGACTG.......................................................................................... | 23 | 0 | 1 | 43.00 | 43 | 2 | 0 | 0 | 1 | 0 | 1 | 38 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGCCCATGGACT........................................................................................... | 24 | 1 | 1 | 43.00 | 43 | 4 | 5 | 10 | 0 | 16 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................CTGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 1 | 1 | 43.00 | 43 | 6 | 8 | 8 | 6 | 9 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCATGGGCT........................................................................................... | 24 | 1 | 1 | 43.00 | 43 | 6 | 10 | 6 | 3 | 10 | 6 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGCGCGAGGAAGTCCATGGACT........................................................................................... | 22 | 2 | 1 | 42.00 | 42 | 2 | 6 | 3 | 0 | 19 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGAACT........................................................................................... | 22 | 1 | 1 | 42.00 | 42 | 8 | 3 | 4 | 8 | 6 | 2 | 9 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAACTCCATGGACT........................................................................................... | 22 | 1 | 1 | 42.00 | 42 | 1 | 6 | 3 | 3 | 4 | 1 | 1 | 1 | 1 | 10 | 2 | 9 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATTAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 42.00 | 42 | 1 | 4 | 7 | 1 | 19 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTTTTCGCGCGATGAAGTCCATGGACT........................................................................................... | 27 | 3 | 1 | 42.00 | 42 | 41 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGATGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 42.00 | 42 | 3 | 1 | 3 | 0 | 5 | 1 | 3 | 16 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATCGACT........................................................................................... | 22 | 1 | 1 | 42.00 | 42 | 1 | 8 | 1 | 1 | 13 | 6 | 6 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAATTCCATGGACT........................................................................................... | 22 | 1 | 1 | 42.00 | 42 | 1 | 10 | 4 | 1 | 8 | 3 | 4 | 0 | 1 | 2 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGTGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 41.00 | 41 | 3 | 11 | 8 | 1 | 9 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGGCCATGGACT........................................................................................... | 22 | 1 | 1 | 41.00 | 41 | 1 | 17 | 14 | 1 | 3 | 2 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCATGGAT............................................................................................ | 21 | 1 | 1 | 41.00 | 41 | 0 | 11 | 6 | 0 | 19 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGACGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 41.00 | 41 | 0 | 1 | 1 | 1 | 6 | 0 | 3 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGAAGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 40.00 | 40 | 3 | 4 | 5 | 3 | 13 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGTTGAAGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 40.00 | 40 | 1 | 8 | 6 | 6 | 2 | 4 | 5 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGTAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 40.00 | 40 | 21 | 0 | 0 | 7 | 3 | 1 | 1 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGAAGTCCGTGGACT........................................................................................... | 22 | 1 | 1 | 39.00 | 39 | 3 | 1 | 4 | 0 | 3 | 2 | 16 | 2 | 5 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGCGCGATGATGTCCATGGACT........................................................................................... | 23 | 1 | 1 | 39.00 | 39 | 3 | 7 | 4 | 8 | 7 | 3 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCAGGGACT........................................................................................... | 24 | 2 | 1 | 39.00 | 39 | 5 | 11 | 12 | 2 | 5 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGACGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 39.00 | 39 | 10 | 6 | 3 | 4 | 9 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................ACAGCGCGCGATGAAGTCCATGGACT........................................................................................... | 26 | 3 | 1 | 39.00 | 39 | 2 | 7 | 6 | 18 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CGCGATGAAGTCCATGGAC............................................................................................ | 19 | 0 | 1 | 39.00 | 39 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 34 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGCGCGATGTAGTCCATGGACT........................................................................................... | 22 | 1 | 1 | 39.00 | 39 | 19 | 2 | 0 | 5 | 3 | 3 | 4 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGGTGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 39.00 | 39 | 3 | 8 | 12 | 2 | 7 | 3 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AGCGCGCGATGAAGTCCATGGA............................................................................................. | 22 | 1 | 1 | 39.00 | 39 | 2 | 5 | 4 | 1 | 10 | 4 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TGCGCGAAGAAGTCCATGGACT........................................................................................... | 22 | 2 | 1 | 39.00 | 39 | 16 | 4 | 0 | 1 | 11 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGGGATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 38.00 | 38 | 3 | 12 | 6 | 0 | 6 | 6 | 1 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCGATGAAATCCATGGACT........................................................................................... | 21 | 1 | 1 | 38.00 | 38 | 6 | 13 | 3 | 0 | 5 | 5 | 2 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................ACCGCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 2 | 1 | 38.00 | 38 | 0 | 3 | 5 | 12 | 4 | 10 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCCCGCGATGAAGTCCATGGACT........................................................................................... | 24 | 1 | 1 | 38.00 | 38 | 5 | 7 | 10 | 1 | 13 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................TGCGCGCGATGAAGTCCACGGACT........................................................................................... | 24 | 1 | 1 | 38.00 | 38 | 7 | 5 | 8 | 3 | 9 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GCGCAATGAAGTCCATGGACT........................................................................................... | 21 | 1 | 1 | 37.00 | 37 | 10 | 3 | 2 | 1 | 11 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................ACGCGCGCGATGAAGTCCATGGACT........................................................................................... | 25 | 2 | 1 | 37.00 | 37 | 8 | 1 | 0 | 16 | 5 | 4 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2l:7179736-7179897 - | dsi_88 | CATTTGGCTT-ATTTAT-AT---------------------GGCGGAGA--------TGG-AG----ATATATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-A-----AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGCCG--GGGGAACATCAT---C----ACATATA---ACGAGTAGG-------- |
| droSec2 | scaffold_3:2941222-2941383 - | CATTTGGCTT-ATTTAT-AT---------------------GACGGAGA--------TGG-AG----ATATATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-A-----AAAAACAAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGTCG--GGGGAACATCAT---C----ACATATA---ACGAGTAGG-------- | |
| dm3 | chr2L:7425762-7425923 - | dme-mir-275-as | CATTTCGCTT-ATTTAT-AT---------------------GGTGAAGA--------TGG-AG----ATATATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-A-----AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--A---------CAGTCG--GGGGAACATCAT---C----ACATATA---ACGAGTAGG-------- |
| droEre2 | scaffold_4929:16344488-16344649 - | CATTTGTCTT-ATTTAT-AT---------------------GGTGGAGA--------TGG-AG----ATATATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-A-----AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGTCG--GGGGAACATCAT---C----ACATACA---ACGAGTAGG-------- | |
| droYak3 | 2L:5473750-5473915 + | CTTTTGGCTT-ATTTAT-AT-------------------ATCGTGGAGT--------TGG-AG----ATATATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-A-----AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTCTGTT---------CAGTCG--GGGGAACATCAT---C----GCATATA---ACGAGTAGG-------- | |
| droEug1 | scf7180000409463:614664-614829 - | TTAA--------------AT-------------------GCGATGAAGA---CATAT-GG-AG----ATGTATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATGT-------AAAAACCAGCCCCGGTCTCTGATTAGCGCGCAAGGTAGAAGACATT--TAAGCCTTTTCA---G--GGAGAACATCAT---TAT--ACATATACTGCCGAGTAGG-------- | |
| droBia1 | scf7180000302422:9125490-9125647 - | TTAA--------------AG-------------------GCGGTGGATG--------TGG-AG----ATGTCTGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAC-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTCT-TT---------AAGCCG--GGGGAACATCAT---C------ATATG---CCGAGTTGGGGCTGTCG | |
| droTak1 | scf7180000415604:142411-142560 + | CGTTT---------------------------------------GGAGATGGGT---TGG-AG----ATGTATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGCCG--GGGGAACATCGT---C----ATATATA---CCGAGTAGG-------- | |
| droEle1 | scf7180000491028:1624838-1624963 + | AA---------------------------------------------AA--------TGA-AG----ATATATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-------GAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGTTG---GGGAACATCAT----------ATGTA-------------------- | |
| droRho1 | scf7180000766421:7650-7778 + | G--------------------------------------------GAAA--------TGA-AG----ATATATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATATGA-----AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--T---------CAGCT---GGGGAACATCAT----------ATATA-------------------- | |
| droFic1 | scf7180000454115:155650-155799 - | CTAA-----------AC-GT---------------------GCTGGATA--------TGG-AG----ATGTATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGCAGTAGACTTT-TT-------TCCAGCT---GGGGAACATCAT----------GTATA---CCGAGTAGG-------- | |
| droKik1 | scf7180000302384:54807-54970 + | CACGAGGTATGAGTTGT--T------------------------GGC-----------GA-TG----ATATATGTCTGG-CACCACGCGCGCTACTTCAGGTACCTGACTGTATAT-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAAGAGACTTT-TT---------CAGCTGGGAAGGAACA-CATCCCGTCCCGCATAAG---CCAAGCTGG-------- | |
| droAna3 | scaffold_12916:15333989-15334108 - | CATTATATAC-TTTTGT-AT---------------------GGC-----------------GG----ATATATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGT-TGC-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTG----------------------GAGA---------------------------------G-------- | |
| droBip1 | scf7180000396572:1689373-1689520 + | TATATACTTT-GCATGT-AT---------------------GTCGGATA--------TGGCGG----ATATATGTCTGC-CACCACGCGCGCTACTTCAGGTACCTGACTGT-TGC-------AAAAACCAGCCCCGGTCACTGATTAGCGCGCAAGGTAGGAGACTTT--G---------GAG---------TACATGG---------------------GAGTTGGAG---TAT | |
| dp5 | 4_group1:4175730-4175850 - | CGAT----------------------------------------G---ATGCCACAC----AGATGT--TCGTGTCTAG-CACCACGCGCGCTACTTCAGGTACCTGACTGCAGTG-A-----AAAAACCAGCCCCGGTCTCTGATTAGCGCGCATGGTAGGAGACTTT---------------------------------------------------CAAGAAGG-------- | |
| droPer2 | scaffold_5:1086439-1086559 + | CGAT----------------------------------------G---ATGCCACAC----AGATGT--TCGTGTCTAG-CACCACGCGCGCTACTTCAGGTACCTGACTGCAGTG-A-----AAAAACCAGCCCCGGTCTCTGATTAGCGCGCATGGTAGGAGACTTT---------------------------------------------------CAAGAAGG-------- | |
| droWil2 | scf2_1100000004585:5257547-5257692 + | AAAATGGTTT-TTTTGTTGTGTCGTCGTTGTCGCCGTCG-TCG-TG---------------CGATGAATCTTGGTCTAA-AACCACGCGCGCTACTTCAGGTACCTGACTGTATTG-AATAAAATTACTCAGCCCTGGTCACTGATTAGCGCGCAAGGTAAGAGAC---------------------------------------------------------------------- | |
| droVir3 | scaffold_12963:15419952-15420067 + | TAAG---C-------------------------------------------------TTA-AGATGA--ATACGTCTGGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGG---GATAACCAAAACCAGCCCCGGTCACTGATTAGCGCGCACGGTAAAAGACGGC--G---------CAGC--------------------------------------------------- | |
| droMoj3 | scaffold_6500:18399373-18399491 + | GCTAGA------ATTGC--T------------------------A------GCAGAG----CGTTGA--ATGCGTCTAGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGA---GATAATCGAAACCAGCCCCGGTCACTGATTAGCGCGCACGGTAAAAGAC---------------------------------------------------------------------- | |
| droGri2 | scaffold_15126:5008401-5008511 - | CAGGCGCCGT-T-------------------------------------------------TGTTGA--ATGCGTCTGGCCACCACGCGCGCTACTTCAGGTACCTGACTGTGG---GATAATCAAAACCAGCCCCGGTCACTGATTAGCGCGCGCGGTAAACGAC---------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 06/28/2015 at 03:54 PM