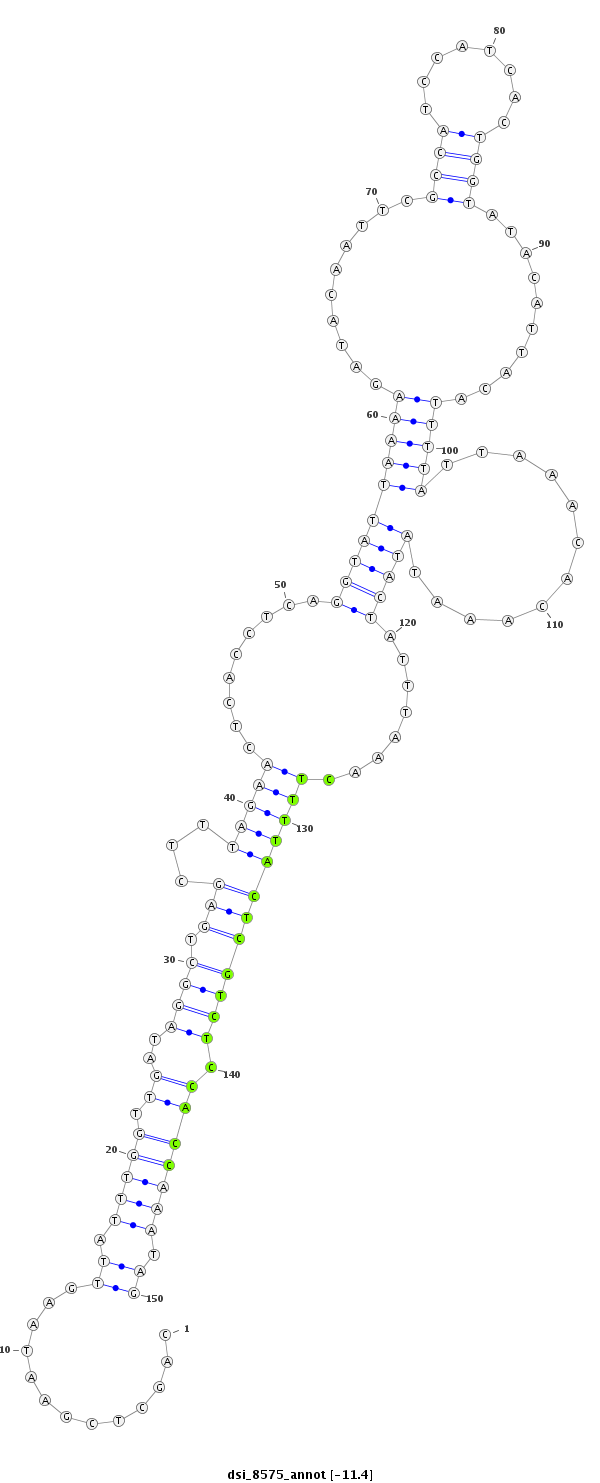
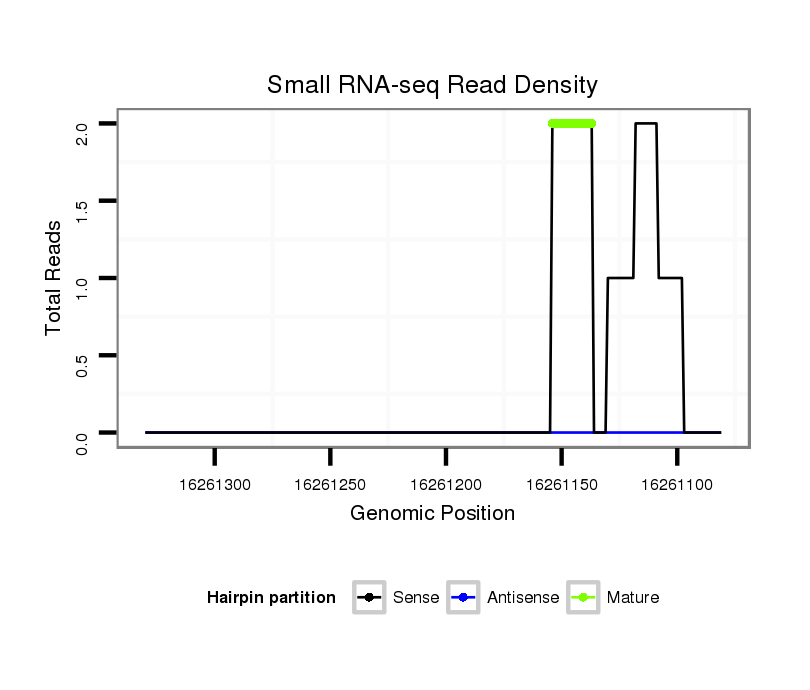
ID:dsi_8575 |
Coordinate:3l:16261131-16261280 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -19.3 | -19.0 | -19.0 | -18.9 |
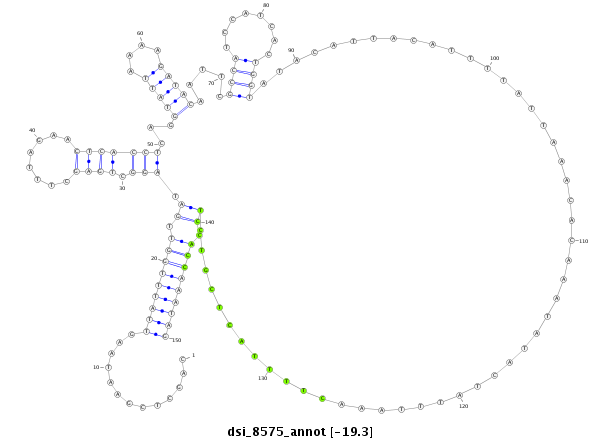 |
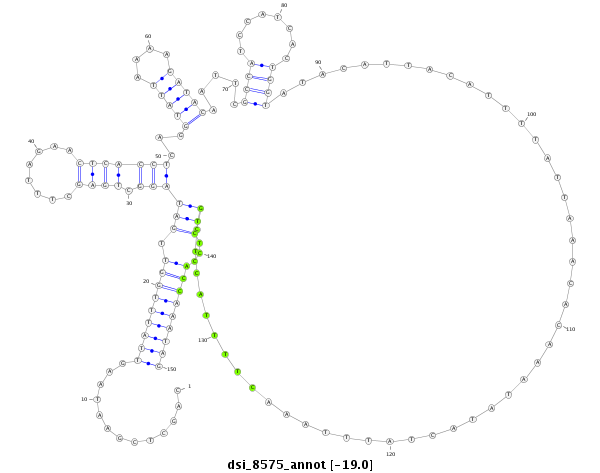 |
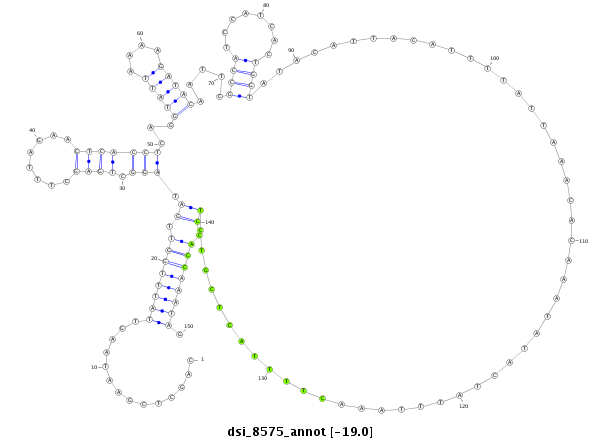 |
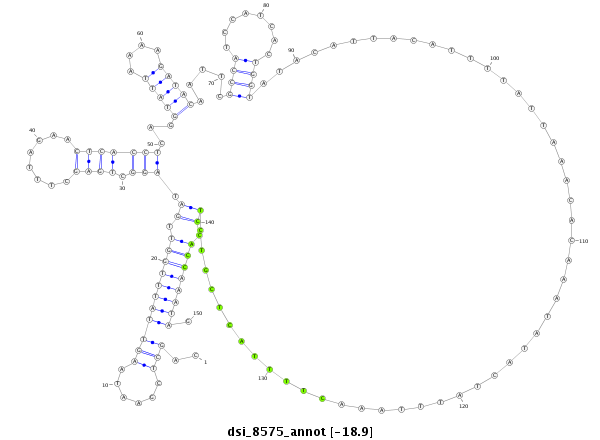 |
exon [3l_16261040_16261130_-]; CDS [3l_16261040_16261130_-]; intron [3l_16261131_16261665_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TAAAATATATTAGGAAACTAATACATACTAGAACTGGCATCTTAGGCCTGCAGCTCGAATAAGTTATTTGGTTGATAGGCTGAGCTTTAGAACTCACCTCAGGTATTAAAAGATACAATTCGCCATCCATCACTGGTATACATTACATTTTATTAAACACAAATATACTATTTAAACTTTTACTCGTCTCCACCAAATAGCATTATTCTGGATGCGGATCCCTCGTCCGTGGTGAAACGCGTTGATCTGC **************************************************.............((.(((((.((..((((.(((...(((((.........((((((((((..........((((........))))..........)))))............)))))........)))))))))))).))))))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M025 embryo |
O002 Head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR1275483 Male prepupae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................CTTTTACTCGTCTCCACC........................................................ | 18 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CATTATTCTGGATGCGGATCCC............................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGCGGATCCCTCGTCCGTGGT................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................GTGGTGAAATGCGTTGA..... | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GGTGAAATGCGTTGAT.... | 16 | 1 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TAGAATATATTAGGAAACT....................................................................................................................................................................................................................................... | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GGTGAAATGCGTTGA..... | 15 | 1 | 11 | 0.45 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CGTCCGTCGTGAAAAGGG......... | 18 | 3 | 20 | 0.40 | 8 | 1 | 0 | 0 | 0 | 0 | 5 | 2 | 0 | 0 |
| ....................................................................................................................................................................................................................................GCGGTGAAATGCGTTGAT.... | 18 | 2 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................GCGCGGTGAAATGCGTTGAT.... | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................TTTGATTGATAGGCTGAGTTTTG................................................................................................................................................................. | 23 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GGTGAAATGCGTAGATCT.. | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CGTCCGTCGTGAAAAG........... | 16 | 2 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................GGTGAAATGCGTTGATAT.. | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GTCGGTGGTGAAACG........... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................................................................GTGAAATGCGTTGATAT.. | 17 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AACTCGTCTCCACCA....................................................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................TATATGGTAGATAGGCT......................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATTTTATATAATCCTTTGATTATGTATGATCTTGACCGTAGAATCCGGACGTCGAGCTTATTCAATAAACCAACTATCCGACTCGAAATCTTGAGTGGAGTCCATAATTTTCTATGTTAAGCGGTAGGTAGTGACCATATGTAATGTAAAATAATTTGTGTTTATATGATAAATTTGAAAATGAGCAGAGGTGGTTTATCGTAATAAGACCTACGCCTAGGGAGCAGGCACCACTTTGCGCAACTAGACG
**************************************************.............((.(((((.((..((((.(((...(((((.........((((((((((..........((((........))))..........)))))............)))))........)))))))))))).))))))).))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................ACGCCTAGGGAGGATGCA.................... | 18 | 2 | 2 | 2.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ..............................................................................TGAGTCGGAATCTTGAGTGGAG...................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................ACGCCTAGGGAGGATGC..................... | 17 | 2 | 12 | 0.42 | 5 | 5 | 0 | 0 | 0 | 0 |
| .............................................................................................................TTTGATGTTAAGCGGTAGG.......................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................ACGCCTAGGGAGGATGCAA................... | 19 | 3 | 15 | 0.20 | 3 | 3 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................ACGCCTAGGGAGCAT....................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....ATATAATGCTTTGATTTTG.................................................................................................................................................................................................................................. | 19 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................ACGCCTAGGGAGGATG...................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 03:39 AM