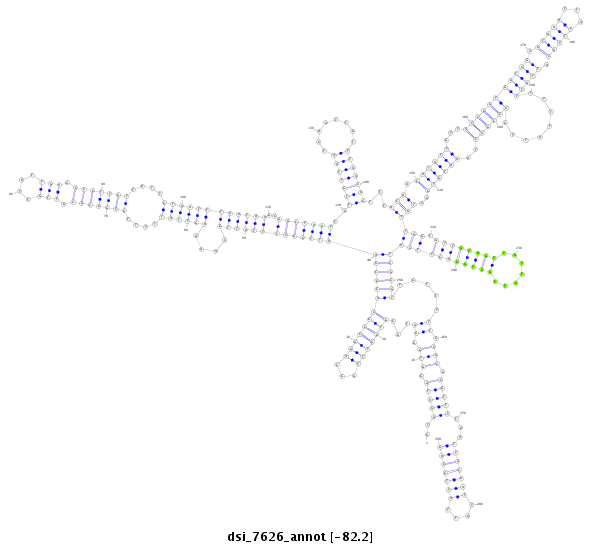
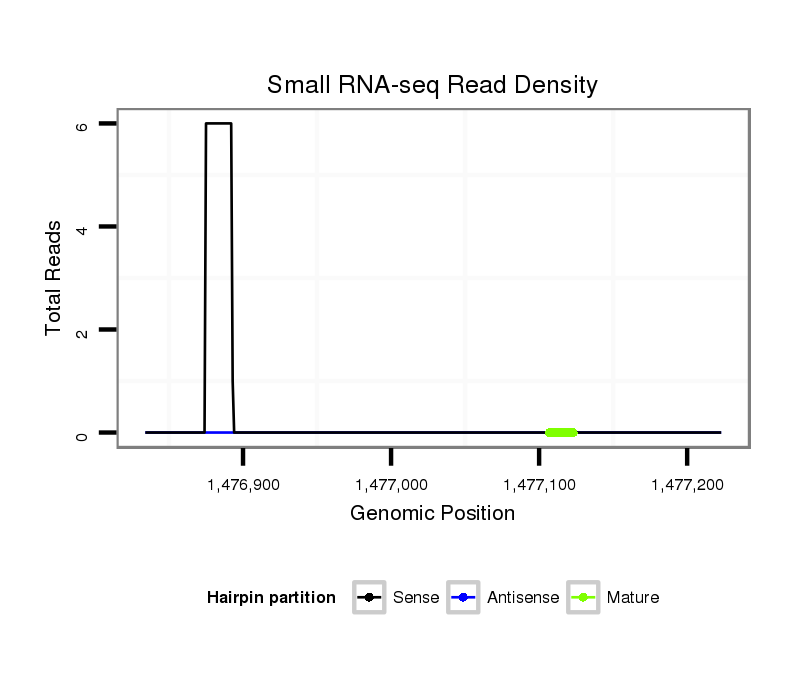
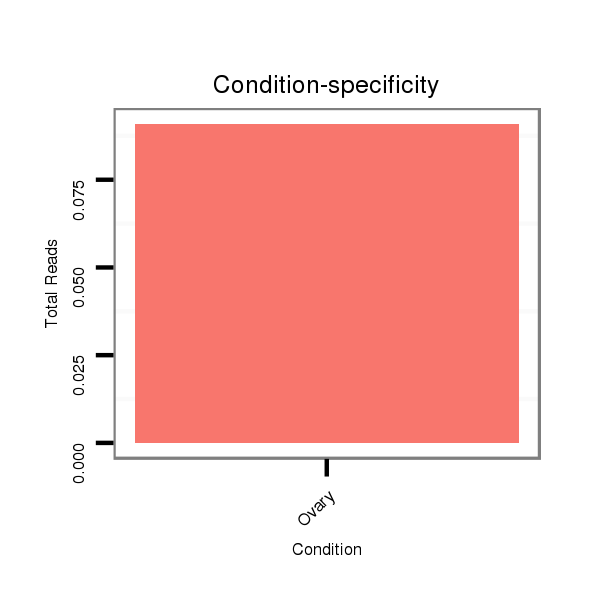
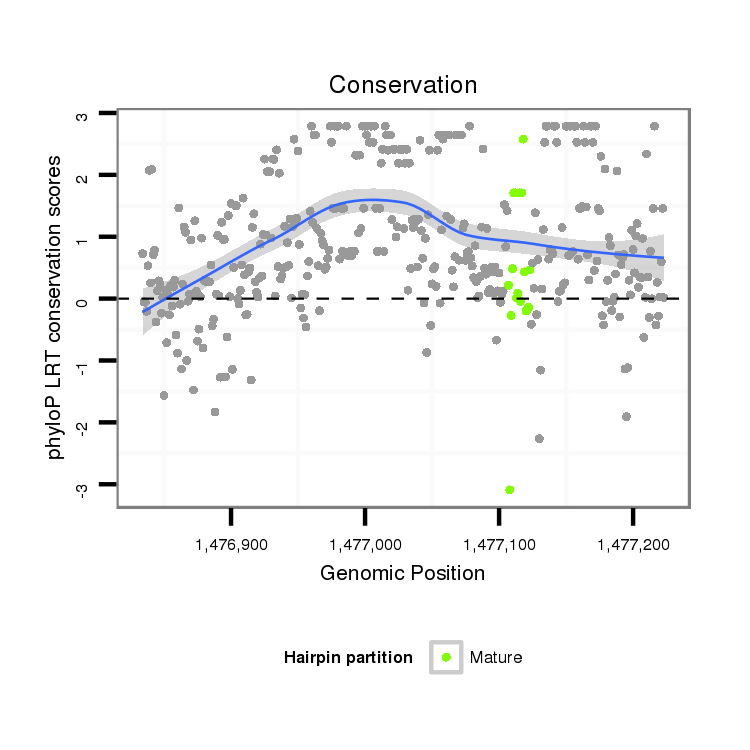
| droSim2 |
x:1476834-1477223 + |
dsi_7626 |
TCAGT-GG-----TGGCAA-------GGTG-GA--------------------------------GGAA-------------TGG---------------ATG----GCTG--GATGG----CTGGATGGATGGGTGGATG----------------------------------------GGTGGGATGG---------GTGGT-------TTG---GCAGGC------CG-CA----------CG--------------CGAATTGGCTT------TATT---GCAAAA--GTTATTTTTGCGGC---GCGTTG------------------------------T--TGTTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGC-------------------------------------C----------------------------GGT----GGTGGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTGGC---------TGTTGGTGTTG---------------TAGTT-GTTGTTGCTGCAGCTCATC---GCG-TTGTTTTTGCA--CGCGTTTTAATTGCAATTGTTAT-------TGCAGTTGGAGCTGCAGTTGC------------------------------------------AGTCCG-CG------------------------------------------------CTGCGT--------------------TTGGT---CTG------------------TTATTACAATT-----A |
| droSec2 |
scaffold_10:1414366-1414740 + |
|
TCAGT-GG-----TGGCAA-------GGTG-----------------------------------------------------------------------------GCTG--GATGG----CTGGATGGATGGGTGGATG----------------------------------------GGTGGGATGG---------GTGGT-------TTG---GCAGGC------GG-CA----------CG--------------CGAATTGGCTT------TATT---GCAAAA--GTTATTTTTGCGGC---GCGTTG------------------------------T--TGTTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGC-------------------------------------C----------------------------GGT----GGTGGTGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTGGC---------TGTTGGTGT------------------TGTT-GTTGTTGCTGCAGCTCAGC---GCG-TTGTTTTTGCA--CGCGTTTTAATTGCAATTGTTAT-------TGCAGTTGGAGCTGCAGTTGC------------------------------------------AGTCCG-CG------------------------------------------------CTGCGT--------------------TTGGT---CTG------------------TTATTACAATT-----A |
| dm3 |
chrX:1609906-1610305 + |
|
TCAGT-GG-----TGGCAA-------GGTG-GA--------------------------------GGAA-------------TGG---------------ATG----GCTG--GATGGGTGGATGAATGA--------CTGGATGGA-TGGGCG---------------------GG--TGGAT-----GG-----GTGG-TGGT-------TTG---GCAGGC------CG-CA----------CG--------------CGAATTGGCTT------TATT---GCAAAA--GTTATTTTTGCGGC---GCGTTG------------------------------T--TGTTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGC-------------------------------------C----------------------------GGT----GGTGGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTGGC---------TCTTGGTGTTG---------------GCGTT-GTTGTTGCTGCAGCTCAGT---GCG-TTGTTTTTGCA--CGCGTTTTAATTGCAATTGTTAT-------TGCAGTTGGAGCTGCAGTTGC------------------------------------------AGTCCG-CG------------------------------------------------CAGCGT--------------------TTGGT---CTG------------------TTATTACAATT-----A |
| droEre2 |
scaffold_4644:1597946-1598350 + |
|
TCAGC-GG-----TGGCAA-------GGTC-GA--------------------------------GGGA-------------TGG---------------GTG----ACTG--GATGG----CTGGTTGGATGGGTGGTTGGGTGGT-TGGGA---------------------------GGTT-----GG-----GAGG------------TTG---GCAGGC------CG-CA----------CG--------------CGAATTGGCTT------TATT---GTAAAA--GTTGTTTTTGCGGC---GCGTTG------------------------------T--TGCTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGC-------------------------------------C----------------------------GGT----GGTGGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTGGC-------AGTGTTGGTGTTG---------------CTGTT-GCTGTTGCAGCAGCTCATC---GCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTGCAGTTGCAGTTGCAG---------------TTGC---------------------AGTCCGAAG------------------------------------------------CTGCGT--------------------TTGTG---CTG------------------TTATTACAATT-----A |
| droYak3 |
X:1462620-1463021 + |
|
TCAGT-GG-----TGGCAA-------GGAT-GA--------------------------------GGAA-------------TGG---------------ATGGGTGGCTG--AACGG----ATGAATGG--------ATGGGAGGA-TGGGAG---------------------GA--TGGGT-----GG-----GTGGTTGGC-------TTG---GCAGGC------CG-CA----------CG--------------CGAGTTGGCTT------TATT---GCAAAA--GTTATTTTTGTGGC---GCGTTG------------------------------T--TGTCGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGC-------------------------------------C----------------------------GGT----GGTGGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCA----G---------------CTAAATGTGGC---------TCTTGGTGTTG---------CTGTTCGTGTT-GTTGTTGCTGCAGCTCAGT---GCG-TTGTTTTTGCA-ACGCATTTTAATTGCAACTGTTAT-------TGCAGTTGGAGTTGCAG------------------------------------------------TCGA-CG------------------------------------------------CTGCGT--------------------TTGTA---CTG------------------TTATTACAATT-----A |
| droEug1 |
scf7180000408994:340715-341141 + |
|
TCAGT-GGCGGCATGGCAA-------GGTCG-GG----AGG-----------------------------------------TGA---------------GTGGGTGGC-------------------------------------------------------------------------CTGGCCTGGTGCCTGGAC-TGGC-------CTG---GCATGTGGC---CG--------TT----------------GTTCGAATTGGCTT------TATT---GCCAAA--GTTATTTTTGTGGCACAGCGTTG------------------------------T--TGTTGAG---GC--------------------ATGGTGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACTGTTTACAACATAT--TGTTGCAAGAACCGTTGTT------GTTGTTGCTG----TTGTC----------------------------GTT----GGTGGCGTAAATTAT-TTGTATTATTTTTTAT-----TGGCCA----G---------------CTAAATGTTGC---------TGT----------------------------------TGGCGCGGCTCAGT---GCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTGCTGCTGCCGTTGCCCC-------------GTTGCAGTTGCATTTGCTGCCATTGCCGCTGC-CA------------------------------------------------TTGCGT--------------------TTGTA---TTG------------------CTATTACAATT-----A |
| droBia1 |
scf7180000301760:4203476-4203889 + |
|
TCAGT-GG-----TGGCAAGCGGCACGGTCGGA--------------------------------GGAG-------------TGG---------------GTA----AGTG--AGTGGTTGGATGGGCGA--------TTGGGCTGA-TGGGCT---------------------GA--TGGGT-----GG-----GTGG-TGGC-------CTG---GCACGC------CGCCA----------TG--------------CGAATTGGCTT------TATT---GCCAAA--GTTATTTTTGCGGC---GCGTTG------------------------------T--TGTTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGTTGCTG---CTGCTGCT------GGTG----------GTGGC----------------------------GGC----GGCGGCGTAAATTAT-TTGTATTATTTTTTAT-----TGGCCG----G---------------CTAAATGCTGT---------TGC---TGT------------------TGTTGGTTTTT-----GGCTCAAC--AGCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTGCAGTTGCAGTCGCAG-----------------------------------------------AG------------------------------------------------CTGCGT--------------------TTGTA---TTG------------------TTATTACAATT-----A |
| droTak1 |
scf7180000415704:1204565-1204894 + |
|
TCAGT-GG-----TGGCAA-------GGTC-GAAG------GTG----------------------GAC-------------TGG---------------GTGGGTGGC--------------------------TTATTG----------------------------------------GGTGGACTAG---------GTGGC-------TCGGCCGCATGC------CG-CA----------TG--------------CGAATTGGCTT------TATT---GCCAAA--GTTATTTCTGCGGC---CCGTTG------------------------------T--TGTCGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATAT--TGCTGCTG---G--------------TCG-----------TAGC----------------------------GGT----GATGGCGTAAATTAT-TTGTATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTTGC---------T-------------------------------------------------------G-TTGCCTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTG-------------------------------------------------------------------------------------------------------------------CGT--------------------TTGTA---TTG------------------TTATTACAATT-----A |
| droEle1 |
scf7180000490566:190675-191011 + |
|
TCACT-GG-----TGGCAA-------GTTTG-GG----AGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCA---GCGTGC------G------GATTTTCATGCGAACGCTGGCGTCAGAATTGGCTT------TATT---GCAAAA--GTTATTTTTGCGGCGCGCTGTTG------------------------------T--TGTTGAGGCTGC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAATAGTTTACATCATAC--TGTTGCTG---TTGTAACAGT--CGGTTG-------------TT----------------------------GCT----GTTGGCGTATATTAT-TTGTATTATTTTTTAT-----TGGCCG----A---------------CTAAATGTTGC---------TGT----------------------------------TGCCGCGGCTCAAT---GCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTG-------------------------------------------------------------------------------------------------------------------CGT--------------------TTG---------------------------TATTATTATT-----G |
| droRho1 |
scf7180000771100:58531-58869 - |
|
TCAGT-GG-----TGGGAA-------GGTTG-GG----AGG-------------------------CAA-------------TGA---------------ATG--------------------------------------------------------------------------------------------------------------------TATGC------T------GATTGGTATGCGAGTGCGAGAGCCCACATTGGCTT------TATT---GCAAAA--GCT-TTTTTGCGAAGCACTGTTG------------------------------T--TGTTGAGGTGGC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAGCATAT--TGTTGCTG---TTGTTACTGT--T-GTTG-------------TT----------------------------GCT----GGTGGCGTG--TTATTTTGTATTATTTTTTAT-----TAGCCG----G---------------CGAAATATTGC---------TGT----------------------------------TGCCGCAGCTCAGT---GCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTG-------------------------------------------------------------------------------------------------------------------CGT--------------------TCG---------------------------TATTGTTATT-----G |
| droFic1 |
scf7180000454072:1486263-1486647 - |
|
TCAGTTGG-----TGGCCA-------GATCG-GA----------------------------------A-------------CTG---------------GTG----ACTG--G----------------------------GCGGAGTGGGCGTGGC---T-------------GG--TGGG------GG-----GTGGTCGGT-------TTG---GCCTGCGGACGACG-CACGAATCG----------------AATCGAATCGGCTT------TATT---GCAAAA--GTTATTTTTGCGGCAC-GCGTTG------------------------------T--TGTTGAG---GC--------------------GC--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACGTAT--TGTTGCGC---TCGCTACAAT--C-GTTG-------------TT----------------------------GCT----GGTGGCGTGAATTAT-TTGTATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTTGC---------CGC----------------------------------TGCCACGGCTCAGT---GCG-TTGTTTTTGCA--CGCATTTTAATTGCAATTGCTAT-------TGCAGTTGCCGTCGCAGTTGCA---------------------------T----------------------------------T----------------------------------------T--------------------GTG---------------------------AATTGTTATT-----G |
| droKik1 |
scf7180000302544:1534539-1534895 + |
|
GG-CT----------GGAA-------GAAG-----------ACGGAGGGGAGTGACCCGGACG---------------------------------------------------------------------------------------GGAC---------------------GA-------------------------------------------GCTC------TG-CA----------TC--------------CGAATTGGCTT------TATT---GCAAAA--GTTATTTTTGTGGC---GCGATG------------------------------T--TGTTGAG---GC--------------------GT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATGT--GGTTGT-------------------------------------TG---CCTACCCCTG--------------CCT----GCCAATGTGAATTAT-TTGCGTTATTTTTTAT-----TG----------------------------------------------------T-------------------------------TACTGTTTGTT---GCGTTTGTTTTTGTA--TGCATTTTAATTGCAATTGTTAT-------TGCAGTTGCAGT------------------------------------------------------TTG-CGATGCATTTGTATTTGTGATTGTGATTGTAATTGTGTTTTTTGTTTTTGTT----------------------------TCTGTTT------------------GTTTTATTACT-----G |
| droAna3 |
scaffold_12929:1493492-1493820 + |
|
CTG--------------------------------------------------------------------------------------------------------------------------AATG---------------------------------------------------------------------------------------------------------T----------CC--------------TGAATTGGCAT------TGTT---GCACAATTGCCATTTT-------------TG------------------------------T--TGTCAAGAG-GC-------------------GGT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTGCAACATGT--TGTTAT-------------------------------------T----------------------------GTTGGTCGGCCGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCCG----G---------------CTAAATGTTGC---------CTT----------------------------------TG---CTTCTTA--CGCCTG-TTGTTTTTGGA--CGCATTTTAATTGCAATTGTTAT-------TGCAGTTGCTGTTGCTGCTGCTGC---------------------TG---CTGCTGCAATTGCCGTTGA-TG------------------------------------------------TTGCAT--------------------TTGTA--TTTGTCATTGTCTTTGTTTTTTGTATTGTTGTT-----G |
| droBip1 |
scf7180000395470:44638-44992 - |
|
TCAG-------------AA-------GG-------------GTGGACT---------------------------------------------------------------------C----CTGAATGA--------G-----------------------------------------------------------------------------------------------T----------CC--------------TGAATTGGCTT------TATT---GCACAATAGT-----------C---GCGTTG------------------------------T--TGTCAAG---GGGTTGGTCGGTCGGTTGGTTGGT--TGTTATTGTAATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTGCAACATGTGTTGTTAT------TGT------------TG-------------TC----------------------------GGTGGTCGGTCGCGTAAATTAT-TTGCATTATTTTTTAT-----TGGCTG----G---------------CTAAATGTTGC---------CTT----------------------------------TT---CTTCTTA--TGCCTG-TTGTTTTTGGA--CGAATTTTAATTGCAATTGTTAT-------TGCAGTTGCTGTTGCAGTTGCAGT-------------GTTGCAATTG---TTGCTGCAATTGC-------TG------------------------------------------------TTGCAT--------------------TTGTG--TTTG------------------TCATTGTCTTT-----G |
| dp5 |
XL_group1a:4366149-4366667 - |
|
TCAGA-GG---------------------------------GCGGACATCGGTCATTGGGGCGTGGTAATGGAGCTGGTGCT-GCGTTTGTGGCGTTGGTATT----TCTGGTTATGG----CTTG------------GGGCGAG-------------TATAGGGTGTACGTTTTGTTA--CATGGCATGG-----GTAT-TGGC------ATTG---GCATGC------G------AGTTT----------------GCACGAATTGGCTTTTTTT----T-------------TGTTTTTGTTTTG---TGTTTTTGTTGTTGCTGTTGTTATTGCTGTTGCTGTTGTGTCGAA---GC--------------------GA--TGTTATTGTGATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATGT--TGTTGCCT---TCGTTGCTGTGGCGG------TTGTTGT----TGTTGT----TGCTGCCTTTGTTGCTGTTGTT----GGTGGCGTGAATTAT-TTGCATTATTTTTTAT-----TGGCTGACT-G---------------CTAAATGTTGA---------TATT---GT----------------------------TG---CTGCTGT--TGCTTG-TTGTTTTTGCACACGCATTTTAATTGCAATTGTTAT-------TGCAGTGGCAGTGGCAGTGGCAGTG---------GCCGTGGC---------------A------GTTGC-AG------------------------------------------------TTGCAT--------------------GTG---------------------------TATTGTTATT-----A |
| droPer2 |
scaffold_11:946594-947099 - |
|
TCAGA-GG---------------------------------GCGGACATCGGTCATTGGGGCGTGGTAATGGAGCTGGTGCT-GCGTTTGTGGCGTTGGTATT----TCTGGTTATGG----CTTG------------GGGCGAG-------------TATTGGGTGTACGTTTTGG-------GGCATGG-----GTAT-TGGCATTGGCATTG---GCATGC------G------AGTTT----------------GCACGAATTGGCTT------TTTT-------------TGTTTTTGTTTTG---TGTTTTTGTTGTTGCGTTTGTTATTGCTGTTGCTGTTGTGTCGAA---GC--------------------GA--TGTTATTGTGATTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAACATGT--TGTTGCCT---TCGTTGCTGTGGCGG------TTGTTGT----TGTTGT----TGCTGCCTTTGTTGCTGTTGTT----GGTGGCGTGAATTAT-TTGCATTATTTTTTAT-----TGGCTGACT-G---------------CTAAATGTTGA---------TATT---GT----------------------------TG---CTGCTGT--TGCTTG-TTGTTTTTGCACACGCATTTTAATTGCAATTGTTAT-------TGCAGTGGCAGTGGCAGTGGCCG---------------TGGC----------------------------AG------------------------------------------------TTGCAT--------------------GTG---------------------------TATTGTTATT-----A |
| droWil2 |
scf2_1100000004515:4072400-4072690 + |
|
TTGTT-GT-----TGTTGT-------TGTT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTT--GTTGTTGTTGTT-------GTTG---TTG------------------------T--TGTTG-----TT--------------------GT--TGTTGTTGTAATTGTTGTT----GTAATTGTTGTTGTT--GTTGTTG------------------------CAAATGTTGTTGTTGTTGTAA------TTGTTGT----T----------------------------GTT----GTTGTTGTA-ATTGT-TGTTGTTGTTGT-TGT-----TG----------------------------------------------------TTGTTGTTGTTGTTGTTGTTGTT-GTTGTTGTTGTTGTTGTTC---TTG-TTGTCGTTGTT--GTTGTTGTTGTTGTAATTGTTGT-------TGTAATTGTTGCTGTAATTGT------------------------------------------TGTTGT-TG------------------------------------------TTGTTGTTGTAA--------------------T--TG---TTG------------------TTGTTGTAATT-----G |
| droVir3 |
scaffold_13042:5185053-5185385 + |
|
TCA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TA---GTAAAG--ATTTATTTTATGGG---CTATTG------------------------------T--TGGC--A---AC--------------------GT--TGTTATTATAACTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAGCTTGT--TATTGCCA---ACGTTGTT------TTTG--------------------------------------------TTGCTTGATGGCGTGAATTAT-TTCCATTTTTGCACATCCATTCAACTGGCTGGAGAGTATATATGTGTTTATGCACTATTGTATTCACTATT---GTTA---------------CTGTT-TTTGTTG----GGCACTGT-AATTG-TTATTTTTGCGCGCTCGTTTTAATTGCAATTGTAATGCGCTCTTGTAGTT------GTAC-----------------------------------------------------------------------------------------------------------ATAT--------------TGTTTGCTG---CTA------------------TTTTTTGGACTGTCCGA |
| droMoj3 |
scaffold_6680:4795965-4796239 - |
|
TTGTT-GT-----TGTTGT-------TACTG-TTACAGTTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------C------------------------------------------------------------------------------------------CAAC---GTGTTGTTGCTGTTACT------------------------------------------------------GT--TGCTATTGCTGTTGCTGTTGCTGCTGA-TGTTGTAGTA--GT-------------------AGT--TGTTGCTG---TTGTTGTTGTCACTG------TTGTTGT----AGTTG-----------------------------------------------------------TTGT-----AGTTTT----A---------------GTAGCTGTTGT---------TGCTGTTGTTG---------------TAGTT-ACTGTTACTGTTGCTATTG---CTG-TTGCTATTGCT--GTTGTTGTCGTTGTTGTTGTAGT-------TGCAGTTGATGCTGTTGCTGT------------------------------------------TATTAG-TG------------------------------------------------CTGT----------------------TGTTG---TTG------------------TTGTTGCTGCT-----G |
| droGri2 |
scaffold_14853:714660-715063 + |
|
TCTGT-GG-----TATCAG-------CATC-AC--------------------------------CG-------------------------------------------------------------------------------------------------------------------------TAG-----GCAT-TGGCTTTGGCATTG---G--------------CT----------TT--------------CGCATTGGCTTTGGCTTTGTCACAGTAAAG--ATTTATTTTATGTC---ATATTG------------------------------T--TA--GCA---GC--------------------AT--TGTTATTATAACTTAATAAGCGGCTAATTGTTGTTAATCAATTGTTTAACAGTTTACAGCTTGT--TATTGCCA---ATTTCGTT------TTTG-----------TTTC----------------------------GCT----GATGGCGTGAATTAT-TTCCATTTTTGCACATCCAT-----------T---------------TTAAGTACCAT---------TGTATTCGC------------------TATT-GTTTTTGTTTCCAGTGT---GTTTG-TTATTTTTGTCCGCTCATTTTAATTGCAATTGTGTTCTGTTTTTGTATAT------ATGTATGTAT--GTATGTATGTATGTAGT----------------------------AC------------------------------------------------TTACATATGTGTGTGTGTGTGT----GTGTG--TGTG----------------TAATTCTCTGCTC-----A |