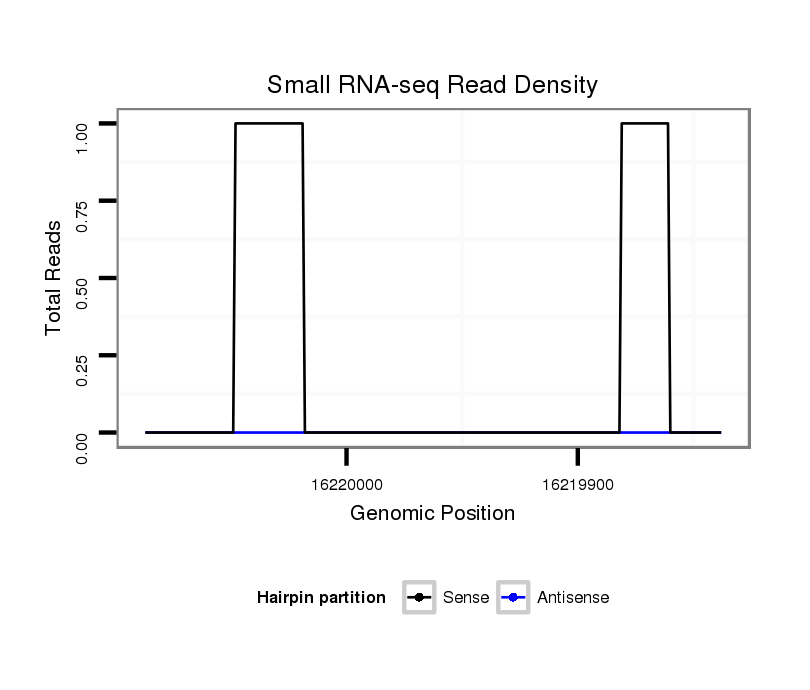
ID:dsi_7367 |
Coordinate:3l:16219888-16220037 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TATTTTGTTTGTAAAAGTTTGTTTCTTTGTTTTGAGCCTCTTCGGTCTGCCATTGTAGTTCATCGCATTTGCCGCCCATCGTCATTAGATGATAAGATGAAAAACTGCCTGCTCTATCCATAGATACTACCAGATTACTAGTTGGCCAGTTGACAACCTGCCTCTCACCCAAAATCTTTCTGTTTCTTTCGTTTCTGCAGGTACCAAGCACAGCTCTTTTCGCGGCCATCCAGGCAAATATCACATGAAC
**************************************************....((((((((((((....)).....((((((....))))))..)))))...)))))((((.........(((.((....(((((.......(((..(((...)))..)))..........)))))....)).))).........))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................TGGCATTTTCCGCACATCGTCAT..................................................................................................................................................................... | 23 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................TGATATGATGAAAAACT................................................................................................................................................ | 17 | 1 | 2 | 1.50 | 3 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................CTTCGGTCTGCCATTGTAGTTCATCGCATT..................................................................................................................................................................................... | 30 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................TAAAACGAAAAACTGCCTGCTCT....................................................................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................AGCACAGCTCTTTTCGCGGCC....................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CAGATTAGTAGTTGG......................................................................................................... | 15 | 1 | 5 | 1.00 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TGGCATTTTCCGCACATCGTCA...................................................................................................................................................................... | 22 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................TGGCATTTTCCGCACATCGTC....................................................................................................................................................................... | 21 | 3 | 7 | 0.29 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................GTTGACAAGCTGCCTC...................................................................................... | 16 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................CGTCTGCCCTTGTATTTCAT........................................................................................................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................GGCATTTTCCGCACATCGTCA...................................................................................................................................................................... | 21 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................TAGATGATTATATGAAAA................................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................GGTAGATCATCGCATT..................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ATAAAACAAACATTTTCAAACAAAGAAACAAAACTCGGAGAAGCCAGACGGTAACATCAAGTAGCGTAAACGGCGGGTAGCAGTAATCTACTATTCTACTTTTTGACGGACGAGATAGGTATCTATGATGGTCTAATGATCAACCGGTCAACTGTTGGACGGAGAGTGGGTTTTAGAAAGACAAAGAAAGCAAAGACGTCCATGGTTCGTGTCGAGAAAAGCGCCGGTAGGTCCGTTTATAGTGTACTTG
**************************************************....((((((((((((....)).....((((((....))))))..)))))...)))))((((.........(((.((....(((((.......(((..(((...)))..)))..........)))))....)).))).........))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .................................CTCTGAGAAGCCAGA.......................................................................................................................................................................................................... | 15 | 1 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................ATGATGGTCTCGTGATCA............................................................................................................ | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................ATTTGACGGGCTAGATAGG................................................................................................................................... | 19 | 3 | 19 | 0.11 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CATGATTCCTGTCGAG.................................. | 16 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................CGGGAGGTCCGTTTA........... | 15 | 1 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................ACGGACGAGATTGGTGACT.............................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................GTGGGTTTCAGACAGAC.................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................ATGCGCTAATGATCA............................................................................................................ | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 03:14 AM