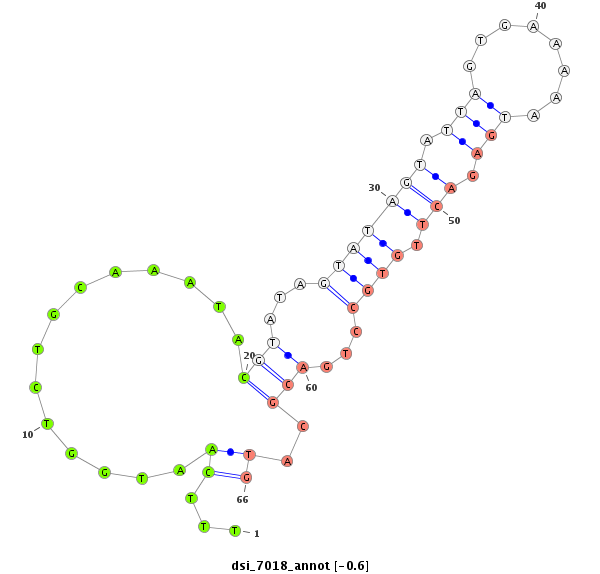
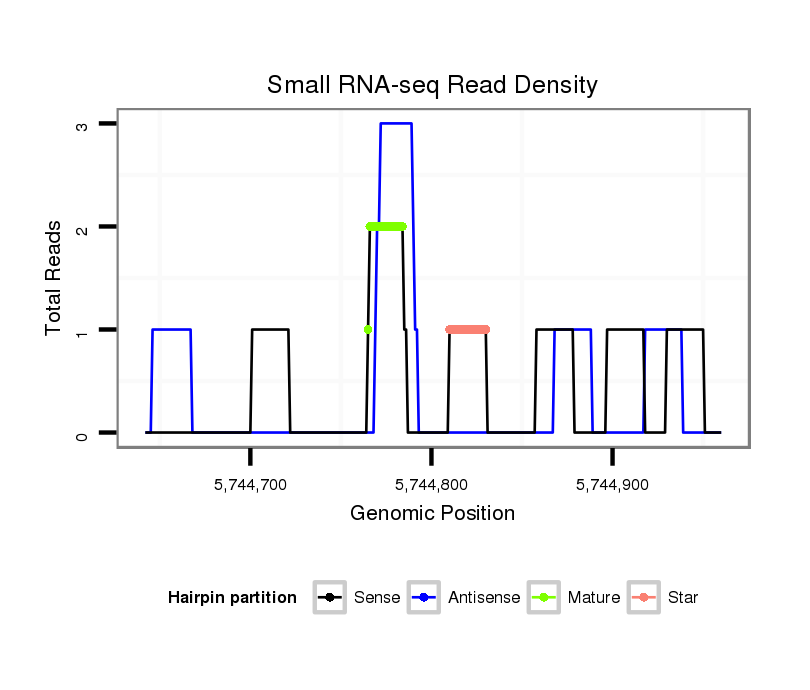
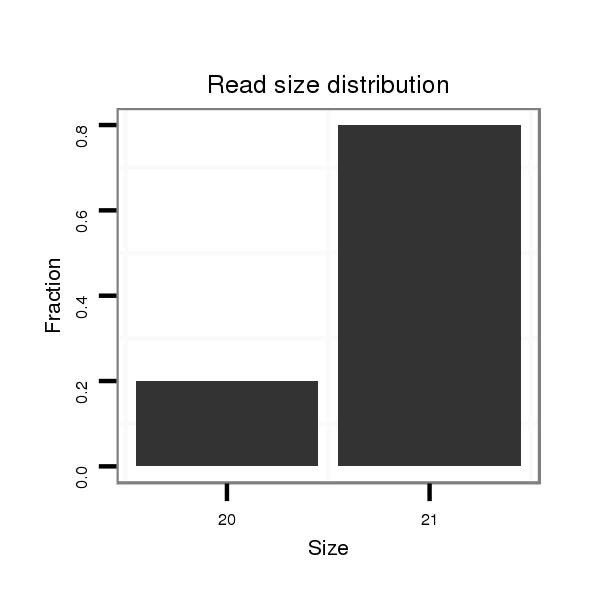
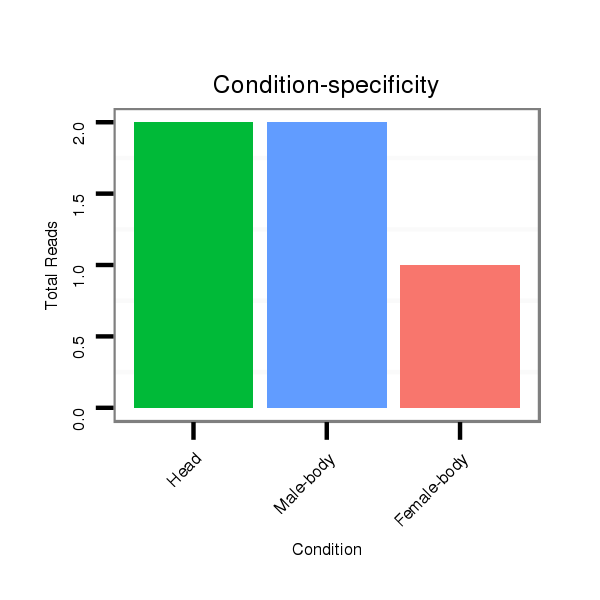
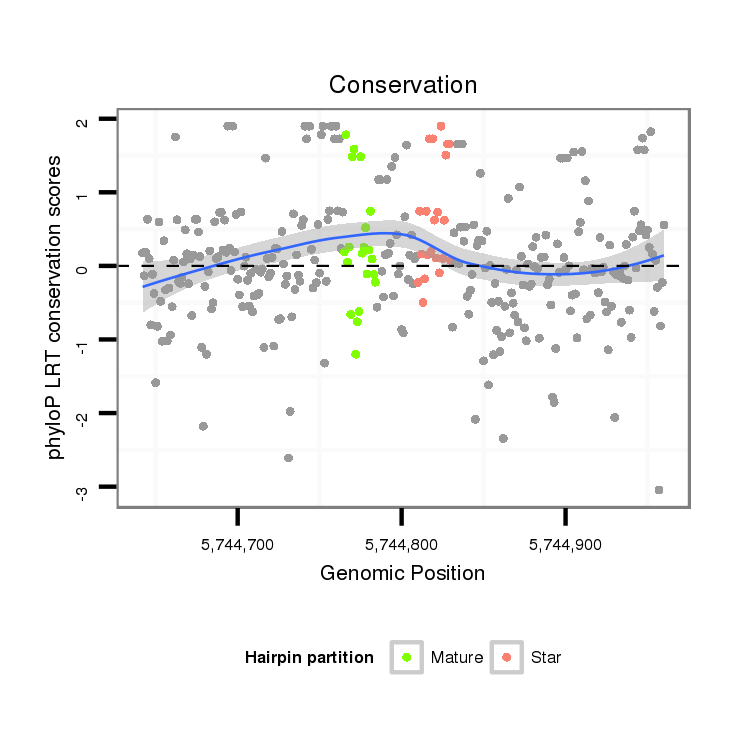
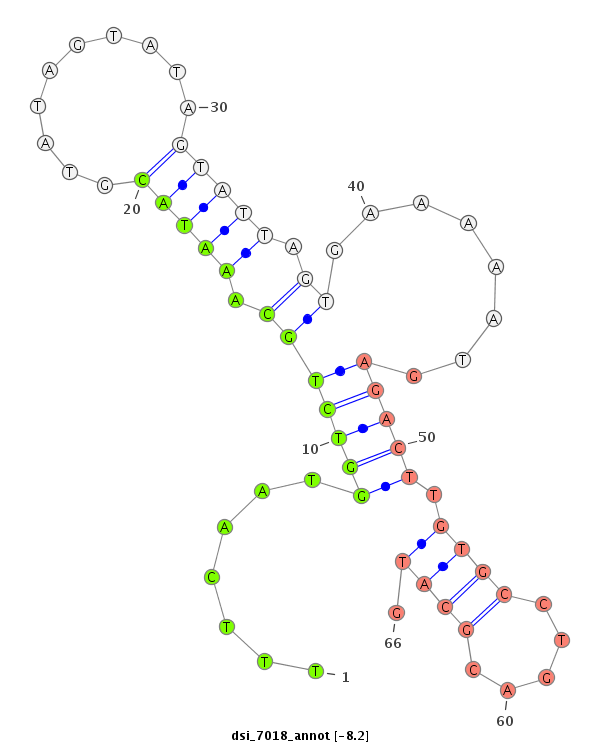
ID:dsi_7018 |
Coordinate:2l:5744692-5744910 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -8.2 | -8.1 | -8.1 | -7.8 |
 |
 |
 |
 |
five_prime_UTR [2l_5744911_5744982_+]; exon [2l_5744911_5745073_+]; five_prime_UTR [2l_5744564_5744691_+]; exon [2l_5744564_5744691_+]; intron [2l_5744692_5744910_+]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TAATGCAGACGACCAACCGGATACATGGTTGAATCAGACCAATCCACAATGTAAATATTAAATGGAAGCTGACGCAGGCAATTTTGCAGCACAGTGGCGTCATACATATTTCTCTTTGTTGTTTTTCAATGGTCTGCAAATACGTATAGTATAGTATTAGTGAAAAATGAGACTTGTGCCTGACGCATGAACTAATCTATTCTAGACACATCCCACCGAGCTGGTGAATCTAAGCCTTGATTACTAAACTGTCATTTCCACTGTTCCAGCACCAAGGAGTCAACCAGAGCACCGGATAAATCGCATGGGATAAAAACAC ***************************************************************************************************************************...((..............(((...(((((((.(((........))).))).))))...)))..))********************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
M053 female body |
O002 Head |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................TTCCACTGTTCCAGCACCAAG........................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TTTCAATGGTCTGCAAATAC................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GAGACTTGTGCCTGACGCATG.................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TTCAATGGTCTGCAAATACGT.............................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................CGAGCTGGTGAATCTAAGCCT.................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................AAATGGAAGCTGACGCAGGCA............................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................GCACCGGATAAATCGCATGGG.......... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................CCCCAGCACCGGATCAATCG................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................CAAGGAGTCAAACAGA............................... | 16 | 1 | 6 | 0.33 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| ......................................................................................................................................................................................................................................................................................................GATGATTCGCATGGGATA....... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................GCATGGGATAGAAAC.. | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................GACACATACCGCCGAGGT................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................ATATTAACTGGAACCTGTC...................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GAATAGTATTAGTGA............................................................................................................................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................AATTTGTCTTTCTTGTTTTTCA............................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
ATTACGTCTGCTGGTTGGCCTATGTACCAACTTAGTCTGGTTAGGTGTTACATTTATAATTTACCTTCGACTGCGTCCGTTAAAACGTCGTGTCACCGCAGTATGTATAAAGAGAAACAACAAAAAGTTACCAGACGTTTATGCATATCATATCATAATCACTTTTTACTCTGAACACGGACTGCGTACTTGATTAGATAAGATCTGTGTAGGGTGGCTCGACCACTTAGATTCGGAACTAATGATTTGACAGTAAAGGTGACAAGGTCGTGGTTCCTCAGTTGGTCTCGTGGCCTATTTAGCGTACCCTATTTTTGTG
**********************************************************************************************************************************...((..............(((...(((((((.(((........))).))).))))...)))..))*************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
O002 Head |
SRR902008 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TTACCAGACGTTTATGCATAT........................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....CGTCTGCTGGTTGGCCTATGTA..................................................................................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................CCAGACGTTTATGCATATCAT........................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TTAGATTCGGAACTAATGATT........................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................CTCAGTTGGTCTCGTGGCCTA...................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TACCAGACGTTTATGCATATC.......................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TCTGCAGTTTGGGCTATGTA..................................................................................................................................................................................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................ACACGGGCGGCGTACTTG............................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................TGGTTGTTCAGTTGGTCACGT............................ | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TCTTCAGTAGGTCTCGTG........................... | 18 | 2 | 9 | 0.22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| .........................................................................................................................AAAAAGTTACTAGACGT..................................................................................................................................................................................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................GTGGTTCGTCAGTTGTTC................................ | 18 | 2 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........TGGTTGGCCTGTGTA..................................................................................................................................................................................................................................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................AGTTGGTCTCGTGTCTTA...................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......TCTGCTCGTAGGCCTAT........................................................................................................................................................................................................................................................................................................ | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TGTAGCTACTTAGTCTGTTT..................................................................................................................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................GTTCCTCTGTTGGTC................................ | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................TTCGAGACTAATGATTTGC..................................................................... | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................TGGGATCGAGGCCTATTT................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................AGGTGGGAAGGTCGTGG.............................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AAAAAAAAGTTACAAGACGTAT................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 2l:5744642-5744960 + | dsi_7018 | TAATG-----CAGACGACCAACCGGATACATG---GTTGAATCAGA--CCAATCCA-CA----ATGTAAATATTAAATGGAAGCTGACGCA------GGCAATTTT-GCAG---------CACAGTGGCGTCATACATA-TTTCTCTTTGTTGTT---TTTCAATGGTCTGCAAATACG------------TATAGTATAGTATTAGTGAAAAATGAGACTTGTGCCTGACGCATG--AA------------CTAATCTATTCTAGACACATCCCACC---GAGC---------------T---G------------------------------------------------------------------GTGAATCTAAGCCT-----TGATTACTA---A---------------------------------ACTGTCATTTCCACTGTT-CCAGCACCAAGGAGTCAAC----CAGAGCACCGGATAAATCGCATGGGATAAAAACAC- |
| droSec2 | scaffold_5:4020243-4020549 + | TAATG-----CAGACGACCAACCGGATACATG---TTTGAATCAGA--CCAATCCA-CA----ATGTAAATATTG-----------ACGCA------GGCAATTTT-GCAG---------AACAGTGGCGTCATACATA-TTTCTCTTTGTTGTT---TTTCAATGGTCTGCAAATACG------------TATAGTATAG-ATTAGTGAAAAATGAGACTTGTGCCTGACGCATG--AA------------CTAATCTATTCTAGACACATCCCACT---GAGC---------------T---G------------------------------------------------------------------GTGCATCTAAGCCT-----TGATTACTA---A---------------------------------ACTGTCATTTCCACTGTT-CCAGCACCAAGGAGTCAAC----CAGAGCACCGAATAAATCGCATGGGATAAAAACAC- | |
| dm3 | chr2L:5949562-5949877 + | TAATG-----CAGACGACCAACTGGATACATG---ATTGAATCCAG--CCGAACCA-CA----ATGTAAATATTAAATGGAAGCTGACGCA------GGCAGTTTT-GCAG---------CACAGTGGCGTCATACATA-TTTCTCTTTGTTGTT---TTTCAATGGTCTGCAAATACG------------TATAGTATAGTATTAGTGAAAAATGAGACTTGTGCCTGACGCATT--AG------------CTAATCTGTCCTGGACACATTACCCC---AAGG---------------T---G------------------------------------------------------------------GGGCATCTAAATCT-----TGATAG-----TA---------------------------------ACAGCCATTT-CACTCTT-TCAGCACCAAGGAGTCATC----CAGAGCACCGAATAAATCGCATGGGATAAAAACAC- | |
| droEre2 | scaffold_4929:6034792-6035108 + | TAATG-----CTGCCGACCATCTGGATACATG---ACTAAATCAGG--CCAAAGCA-CA----ATGTAAATATTAAATGGGATATAATGCA------GGCGATTTC-GCAG---------CGCAGTGGCGTCATACATA-TTTCTCTTTGTTGTT---TTTCTATGGACTGCAAATGCG------------TATAGTATAGTATTAGTGAAAAATGAGTCTTGTGCCTGAGGCATT--AG------------CTAATCTATCCTGGACAATT-TCACT---GGTC---------------T---G------------------------------------------------------------------GTTCATCTAAGCCT-----TGGTAG---AGTA---------------------------------ACA-CCATTT-CACTGTTCCCAGCACCAACAAGTCATC----CAGCACAAGGAATAAATCGCATGGGATAAAAGCAC- | |
| droYak3 | 2L:13246611-13246917 - | TAATG-----CTGTCGACCATCTGGATACATG---ACTAAATCAGG--CCAAACCA-CA----ATGTAAATATTAAATGGGATCTAACGTA------GTCGAACTC-GCAG---------CGCAGTGTCGTCATACATA-TTTCTCTTTGTTGTT---TTTCGATCGACTGCGAATACG------------TATAGTATACTAGTATTGAAAAATGAGACTTGTGCCTGAGGCATT--AG------------CTAATCTATCCTGGACA-------------GGC---------------T---G------------------------------------------------------------------GTGCATCTACACCT-----AGATAG-----TA---------------------------------ACACCTATTT-CACTGTTCCCAGCACCGACGCGTCATC----CAGAGCAACGAATAAATCGCATGGGATAGAAGCAC- | |
| droEug1 | scf7180000409004:15516-15823 - | TAATG-----GCGTTGGACCTATAGATATGTG---ACAGTATCAATTGTGGGTTAG-CA----ATGTAAGTATTGAACGCGCCTTGTCGCA------GGTATTTCC-GCTG---------AACAGTGGCATCATAAATA-TTTTCCTTTGTTGAT---TTTCGATGGACTGCTAATA-----------------------ATATAGATGGAAAAACAGAAATGTGCCTAACGCATT--TG------------TTAATCTGCGTTAGACACATACAAAT---GTA---------------TCTCAT------------------------------------------------------------------------GTAAATGTTCACCTGATAG-----TA---------------------------------ACTAGAATTT-TACAGTC-GCAGCAGCTACGATTATCA----TAGAACAGGGA-AATATCGCATGGGATAAGGACAC- | |
| droBia1 | scf7180000301629:45155-45205 + | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTT-CCAGCA-------GTTACTCAAAACAAGTAACAGGTATAACGCATGGAATA---ACAC- | |
| droTak1 | scf7180000415103:440957-441326 - | TAATG-----CTGACGTT--TCTAGATGTTTG---ACTAATCCAAG--CGAAACCACCAATGTATGTAAATGTGAAACGGGCGTTAACAAA------GGTACTTCT-GCAG---------TGTAGTAATGTCATTCGAGATTTGTTATTGTCGAT---CT-------GCTGCAAATAT--------------------------------------AGAGTTGAGCCTAACGCATTTTTG------------CTAATCTGCCTTGGGCATATACCACCTTTATAATGGGGTCTTAAACAAGTCGCTTAGAGCTAAATTTGGGTACAATTTATATTGATGTAAAAAATAATTTAGTTCGAAATCAGGTTGAAGAGATTCTATGGCA-----CCGCCA---AATA---------------------------------ACTAATATTT-CACAGTC-GCTGCA-----GTGTCACT----CAGAACAAGGAATTTATCGCATGGGATAAA---AC- | |
| droEle1 | scf7180000491120:15300-15588 - | TAATG-----CTATACACCTTCTGGACATACACACAGTCTATCCAG--CCAAGGCA-AA----ATGAATTTATAAAACGGGCGTTATCGCA------GGTATCTC-----A---------AGAAGTGGCGTCATAGATA-TTTTCCTTTGTTGTTTT-TTTCGATAAACAGCAATTT---------------------TATTTGCTGTGAAACTAGAGGCTTGTGCCTAACGCATT--CG------------CTGATCTGTTTTTGACATATACCTCT---GCGC---------------TT--T------------------------------------------------------------------GTG---------------------------TA---------------------------------TTTCGTACTT-TACAGTC-GCAGCACC---GAGCCATC----TAAATCGAGGAACACATCGCATGGGATAAAACTAC- | |
| droRho1 | scf7180000761581:139-435 + | TATTG-----CTATACACCTACCGTATATACATATAGTAAATCCAA--CTAAGCCA-CA----ATGTACATATAAGGCAGACGTTATCGAA------GGTATTT-----AA---------GATAATGGCGTCATAAATA-TTTCCCTTTGTTGTT---TT--GATAGACTGCAAATA---------------------TATTAGTTATGAAAATAGTGACTTGTGCCTAACGCATT-TCG------------ATGATCTGTCTTCGACTCATACATCT---GCGG---------------T---T------------------------------------------------------------------GTGAG--TGAGCCT-----TGATAA-----TA---------------------------------GCTAATGTTT-CACTTTC-GCAGC------GAGTCATT----CAGAGCAAGGAATACATTGGATGGGATAGAAGTGC- | |
| droFic1 | scf7180000453813:98925-99219 - | TAATG-----CTTGTGAGCTGTAGAATATATG---ACTAAATCAGA--CCAAACCG-CA----ACGTAAGTATCAACCGGACGTAATCGCA------GATATTTTC-ACAG---------GGTAGTA--GTCATACAAA-A-TCTCTTTGTTGTT---T-TCGATGCGCTGCATATACAAATGTACATACATA-TGTATATTAGTTGCGAAAATAAAGACTTGTGCCTAACGCATT-TTG------------CTGATCTGCCCC-----------C---TCATAT-------------------T------------------------------------------------------------------GAAATCGTAAGCCC-----TGATAA-----TA---------------------------------G-------TT-CAATATTTGCAGGAACATCGATTCTT---------------AAACAATCGCATGGGATAAAAATAA- | |
| droKik1 | scf7180000302382:578689-578912 - | CAATG-----ATCCCGCT--GCCG--------------------CT--GCAAACCA-CA----GAATAAATATAGAGCTGG----------------TGCG-TCAATATGACGTG-----CCTAACGTCGTCATGCATA-TTTTTCTTTGTTGTC---TTCC-A-CGGCTGCAGACAT-------------TAGTGTATATTAGTGTTGAGACTCGAATCTGGTGCACAACGCATT--TGCCCGAAAAA--TCTAGTTTTACCTC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGG------AATTCATT----TAATTTCAGTAATTCATCGCATGGGA---------- | |
| droAna3 | scaffold_12916:11362436-11362653 + | TGC-----------AGACTCATTAGATGCACG---ACTAAATCAGC--CTGTGCCA-CA----ATGTAAATACAAATTTTG----------------GGC-GTTTC-ACAGCT-------GACTGCAGCGTCATACGCA-GTTGCCTTTGTTGTT---CTGG-----------AATTCC------------TATGG---------CGTGGAATTAGAATCTGGAGCATTACGCATC--CGAGCCAAACTTCTCTAATTATTCCTGAACATTTTCA--------------------------------------------------------------------------------------------------------------------GATAG-----GG---------------------------------A--------------------------------------------------------TTTGCATG--CTGGAGCCACA | |
| droBip1 | scf7180000396572:1029319-1029555 + | TTATG-----ATGCAGACTCATTAGATGCATG---ACTAAATCAGC--CTGTGCCA-TA----ATGTAAATACAAATTTT---------------GGGGCA-TTTCAACGGCT-------GGCTATAGCGTCATACGCA-GTTGCCTTTGTTGTT---TTGGAATTCA-TCCGAAC-------------------ATGCATTAGTCGTGGACATAGAAACTGGTGCATTACGCATC--AGAAGCACAATGCTCTAATTATTCCTGAACATTTGCA--------------------------------------------------------------------------------------------------------------------GATAA-----GGACTTGGA--------------------------------------------------------------------------------------GCGTG--TTG---GAGC- | |
| dp5 | 4_group1:343157-343436 - | TCATCAAAAAGCGCAGCTTCATACGATCGGTG---ATGCAATAAAA--GAAAATC--CT----ATGGAAATCTATAATTAAAACGGAAAAATGTTGGTTTA-TTATTAAGACCTGCCACCACAAATGGCGTCATA-----ATTCTCTTTGTTGTTGTTTTTG--TGCACCCAAAACA--TACG------------------------TGCT-AAAAAGATTTGTGTCTGAAA-------------------------------------ATACCCGAT---GCA---------------TATTATATGGAAATGACTT-----------------------------------------------------------------------------------------GGGGAAAAATGAATATCTAAAGAATATCAAAACAAAATT-CAAAGTC-ACAGC-----------------------------------------TGATTTGTACCC- | |
| droPer2 | scaffold_1:338632-338904 - | AAAAA-----CCGCAGCTTCATACGATCGGTG---ATGCAATAAAA--GAAAATC--CT----ATGGAAATCTATAATTAAAACGGAAAAATGTTGGTTTA-TTATTAAGACCTGCCACCACAAATGGCGTCATA-----ATTCTCTTTGTTGTTGTTTTTG--TGCACCCAAAACA--TACG------------------------TGCT-AAAAAGATTTGTGTCTGAAA-------------------------------------ATACCCGAT---GCA---------------TATTATATGGAGATGACTT-----------------------------------------------------------------------------------------GGGGAAAAATGAATATCTAAAGAATATCAAAACAAAATT-CAAAGTC-ACAGC------------------------------TGATTT-------------GTAC- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/19/2015 at 05:42 AM