
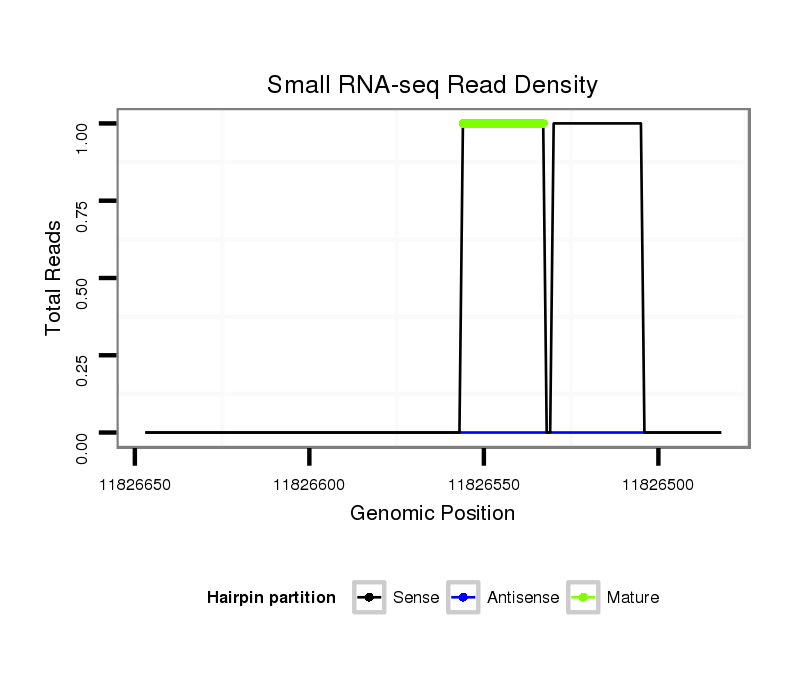
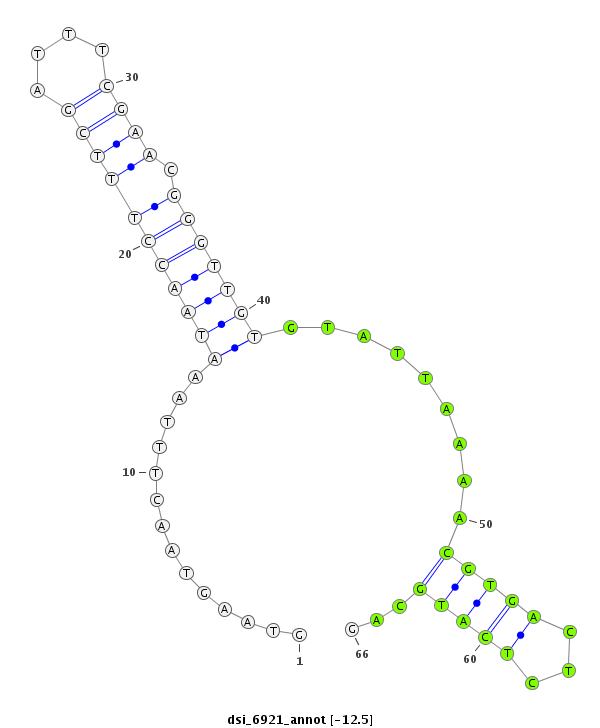
ID:dsi_6921 |
Coordinate:3l:11826532-11826597 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
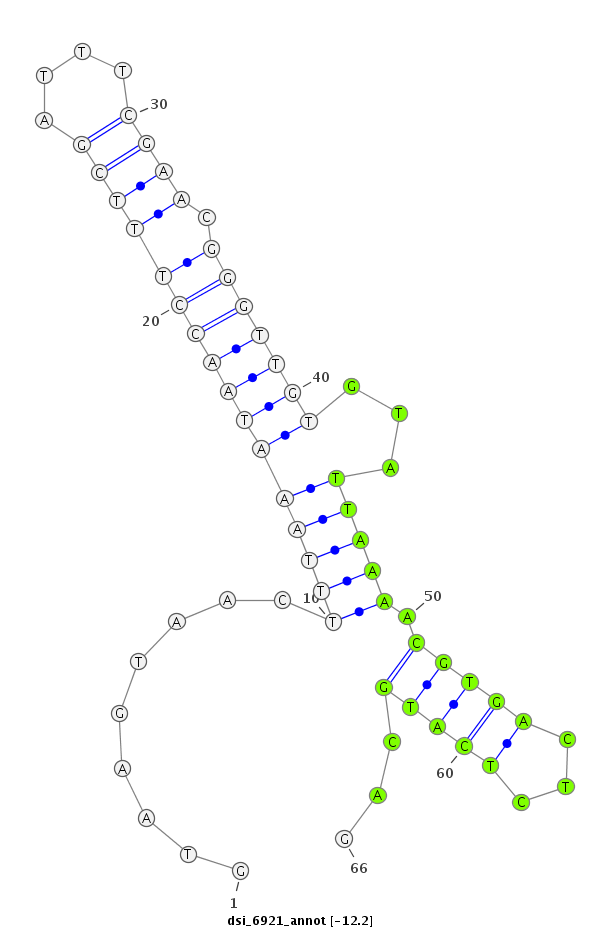
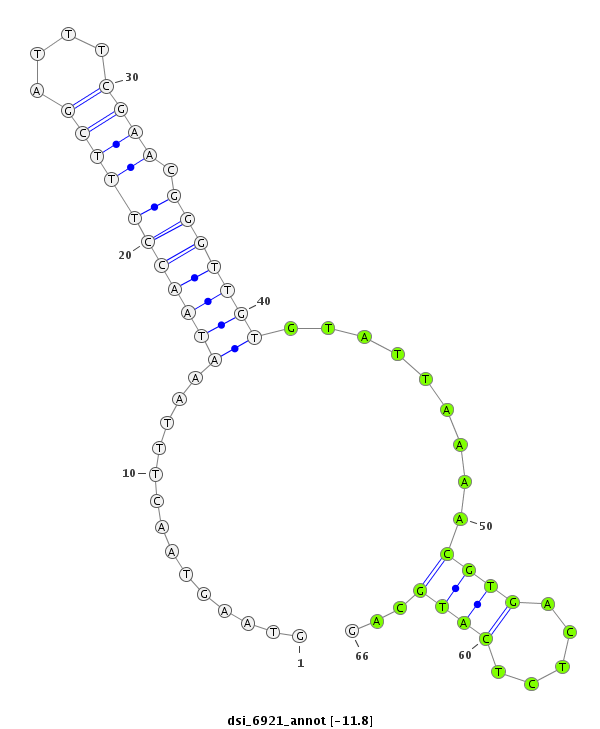
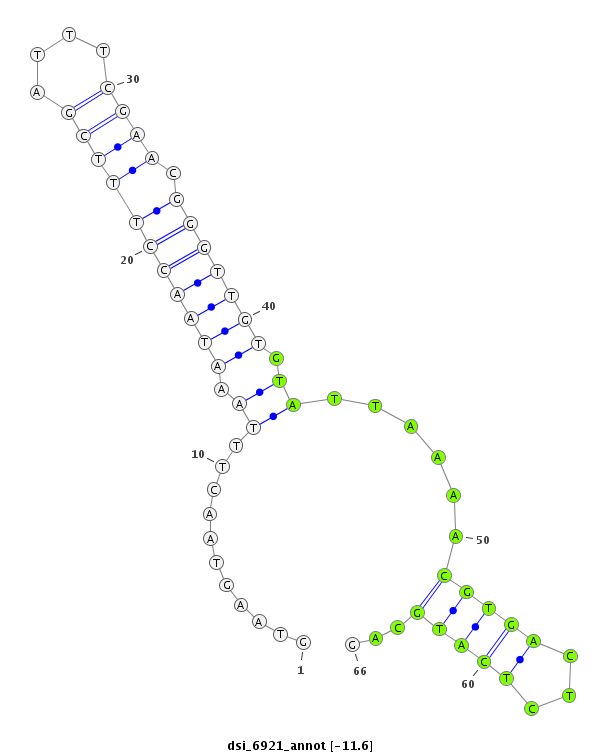
| -12.5 | -12.2 | -11.8 | -11.6 |
 |
 |
 |
 |
CDS [3l_11825745_11826531_-]; exon [3l_11825638_11826531_-]; CDS [3l_11826598_11826761_-]; exon [3l_11826598_11826761_-]; intron [3l_11826532_11826597_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------------################################################## AAGAATGAAGACCTAAAGCGAAAAATATTCAAGAGTCTTTATGGAGAAGGGTAAGTAACTTTAAATAACCTTTCGATTTCGAACGGGTTGTGTATTAAAACGTGACTCTCATGCAGTTCTGACGATGAGGAGCAGCCAGCAGAAGAGAAACCAGCAGTGACACCAG **************************************************........((....(((((((((((....)))).))))))).......................))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M025 embryo |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................TCTGACGATGAGGAGCAGCCAGCAGA....................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GTTCTGACGCTGAGGCGC................................. | 18 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GTATTAAAACGTGACTCTCATGCA................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GTTCTGACGCTGAGGA................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GTTCTGACGCTGAGG.................................... | 15 | 1 | 6 | 0.50 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................CTGACGCTGAGGAGC................................. | 15 | 1 | 8 | 0.50 | 4 | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................CGGTTCTGACGCTGAGGCGC................................. | 20 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TTGATTTCGAACGGG............................................................................... | 15 | 1 | 12 | 0.25 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TCTCATCCAGTTCTGATG.......................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................TTTATTGAGAAGGGT.................................................................................................................. | 15 | 1 | 12 | 0.17 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................AGCACCCAGCACAAGAGA.................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGTTTTAAAACGTGA............................................................. | 16 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................TCGATCTCGAACGGG............................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................TTCTGACGCTGAGGCGC................................. | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................CAGTTCTGACGATGAGCCC.................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................TCCTGACGCTGAGGAGC................................. | 17 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTCTTACTTCTGGATTTCGCTTTTTATAAGTTCTCAGAAATACCTCTTCCCATTCATTGAAATTTATTGGAAAGCTAAAGCTTGCCCAACACATAATTTTGCACTGAGAGTACGTCAAGACTGCTACTCCTCGTCGGTCGTCTTCTCTTTGGTCGTCACTGTGGTC
**************************************************........((....(((((((((((....)))).))))))).......................))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M023 head |
M024 male body |
O002 Head |
M025 embryo |
SRR618934 dsim w501 ovaries |
M053 female body |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................AGGACGTCAAGTGTGCTAC....................................... | 19 | 3 | 20 | 128.85 | 2577 | 686 | 804 | 542 | 196 | 335 | 4 | 0 | 6 | 2 | 0 | 1 | 1 |
| ............................................................................................................AGGACGTCAAGTGTGCTACT...................................... | 20 | 3 | 5 | 115.60 | 578 | 245 | 145 | 75 | 49 | 26 | 2 | 30 | 0 | 4 | 2 | 0 | 0 |
| .............................................................................................................GGACGTCAAGTGTGCTACT...................................... | 19 | 3 | 18 | 19.44 | 350 | 196 | 1 | 1 | 98 | 0 | 33 | 17 | 0 | 4 | 0 | 0 | 0 |
| ...............................................................................................................ACGTCAAGTGTGCTACT...................................... | 17 | 2 | 20 | 12.50 | 250 | 0 | 0 | 0 | 0 | 0 | 236 | 0 | 0 | 1 | 8 | 5 | 0 |
| ............................................................................................................AGGACGTCAAGTGTGCTACTC..................................... | 21 | 3 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ACAGTACGTCAAGTGTGCTAC....................................... | 21 | 3 | 2 | 2.00 | 4 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ACAGGACGTCAAGTCTGCTAC....................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGGACGTCAAGTGTGCTACTCC.................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ACAGTACGTCAAGTGTGCTACT...................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGGACGTCAAGAGTGCTACTGC.................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGTACGTCAAGTGTGCTACTGC.................................... | 22 | 3 | 3 | 0.67 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGTCAAGTCTGCTACTGC.................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGGACGTCAAGTCTGCTAC....................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CGTCAAGTCTGCTACTGC.................................... | 18 | 2 | 4 | 0.50 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................AGAGGACGTCAAGTGTGCTACT...................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGTCAAGTGTGCTACTGCT................................... | 20 | 3 | 7 | 0.43 | 3 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CAGTACGTCAAGTGTGCTAC....................................... | 20 | 3 | 5 | 0.40 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGTCAAGAGTGCTACTG..................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GGACGTCAAGAGTGCTACTG..................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGTACGTCAAGTGTGCTACTG..................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TACGTCAAGTGTGCTACT...................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................GGAAAGTTAAAGCTTG.................................................................................. | 16 | 1 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...............................................................................................................ACGTCAAGTGTGCTACTC..................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................GCCTTTTATAAGTACTCATAA............................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................GTACGTCAAGTGTGCTAC....................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CGTCAAGTCTGCTACTG..................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGCACGTCAAGTGTGCTAC....................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................ACGTCAAGTGTGCTACTCG.................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................ATTGGAAAGCTCATGC..................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........GGATTTCGCATTTTAAA.......................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................TTATTGTCATGCTAAAGCTT................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/24/2015 at 10:38 AM