
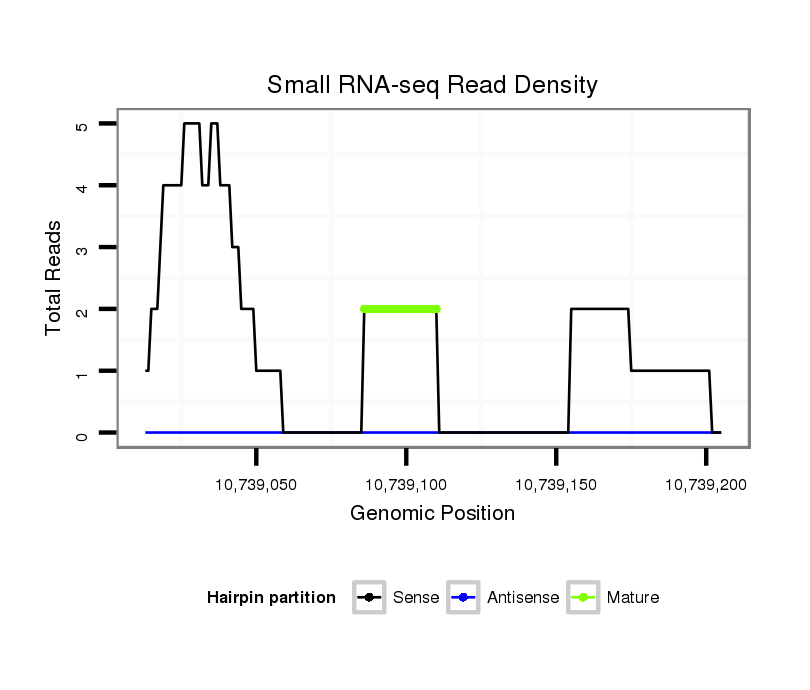

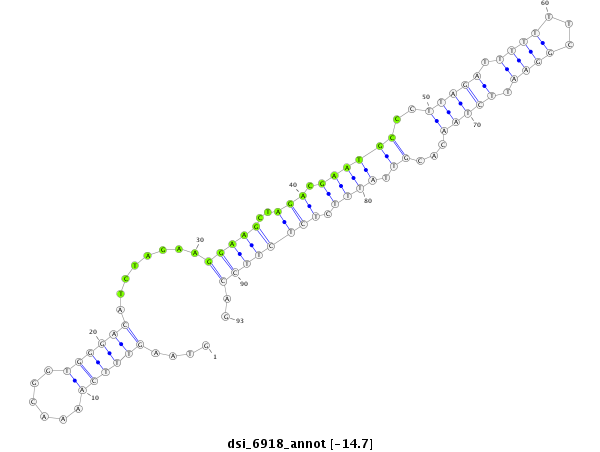
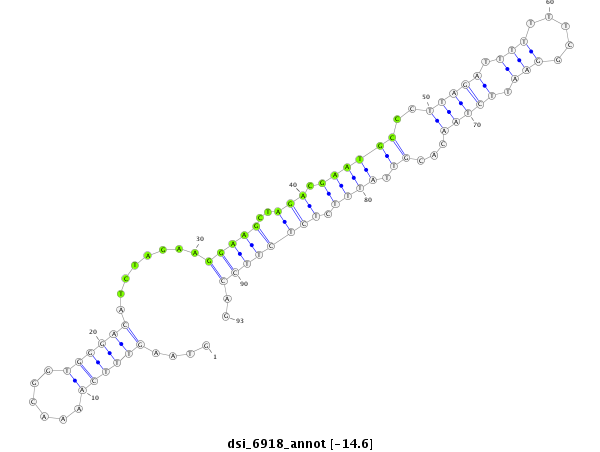
ID:dsi_6918 |
Coordinate:3r:10739063-10739155 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -15.5 | -15.4 | -14.7 | -14.6 |
 |
 |
 |
 |
exon [3r_10737921_10739062_+]; CDS [3r_10737921_10739062_+]; CDS [3r_10739156_10739257_+]; exon [3r_10739156_10739257_+]; intron [3r_10739063_10739155_+]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------------------------------################################################## CGGACTGGGCGGTCTCAGCGAGTCGGAGCTGATAGGCGCTAACGATGCTGGTAAGTTTCAAAACGGTGGGACATCTAGAAGGAAGCTAGACGAATGCCCTTAGATTTTTTTCGGAATTCTAACACGTTATTTCTCTCTTCCAGATGATATAGCATTGAATGGCGCCCACTTGCTGGAGGAGCACGATCTGGTA **************************************************........................((....(((((..(((..((((....((((.(((.....))).))))...))))...)))..)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
O002 Head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TCTAGAAGGAAGCTAGACGAATGCC............................................................................................... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AGTCAGAGCTGATAGGCGCTA........................................................................................................................................................ | 21 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GATGATATAGCATTGAATGG............................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................GATGATATAGCATTGAATGGC.............................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CGGACTGGGCGGTCTCAGC.............................................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............CTCAGCGAGTCGGAGCTGATAGGC............................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TTTTTCGGAATTTTAACAC.................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCGGAGCTGATAGGCGCTAACGAT................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GCCCACTTGCTGGAGGAGCACGATCT.... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GACTGGGCGGTCTCAGCGAGTCGGAGC.................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GGGCGGTCTCAGCGAGTCG........................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....TGGGCGGTCTCAGCGAGTCGGAGCTGA................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........GCGCTGTCGGCGAGTCGG....................................................................................................................................................................... | 18 | 3 | 20 | 0.35 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 1 |
| .......GGCGCTGTCGGCGAGTCGG....................................................................................................................................................................... | 19 | 3 | 20 | 0.35 | 7 | 0 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .TGACTGGGCGGTCTC................................................................................................................................................................................. | 15 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......GGCGCTGTCAGCGAGTCG........................................................................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................TAAAGATGCTGGTAAGAT........................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TCAGCACGGTCGGACATC...................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
GCCTGACCCGCCAGAGTCGCTCAGCCTCGACTATCCGCGATTGCTACGACCATTCAAAGTTTTGCCACCCTGTAGATCTTCCTTCGATCTGCTTACGGGAATCTAAAAAAAGCCTTAAGATTGTGCAATAAAGAGAGAAGGTCTACTATATCGTAACTTACCGCGGGTGAACGACCTCCTCGTGCTAGACCAT
**************************************************........................((....(((((..(((..((((....((((.(((.....))).))))...))))...)))..)))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................AGAGAGAAGGTCTATT.............................................. | 16 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..CTGACCCGCCAGAGG................................................................................................................................................................................ | 15 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| ..........................TCGGCTCGCCGCGATTGCTA................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ............AGAGTCGCTTAGCCT...................................................................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:47 AM