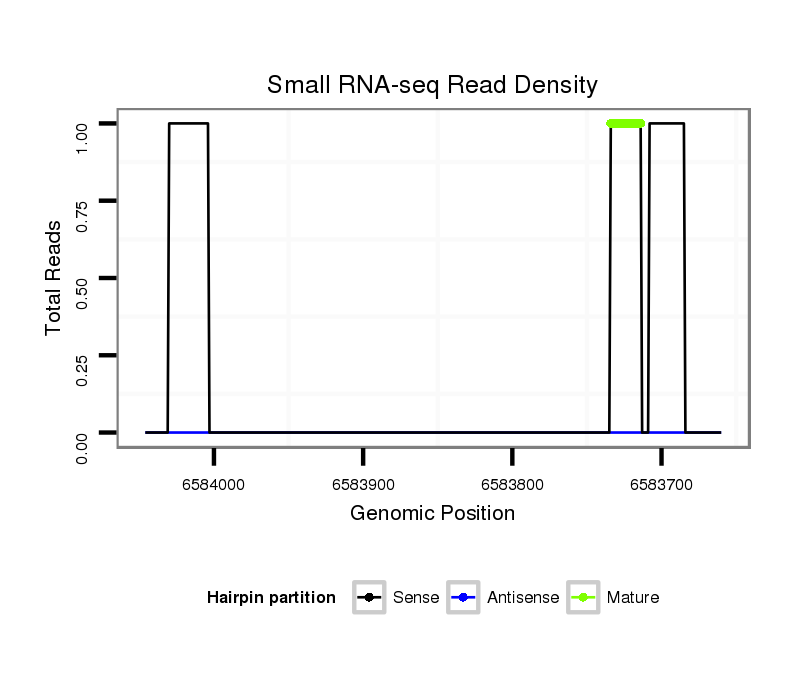
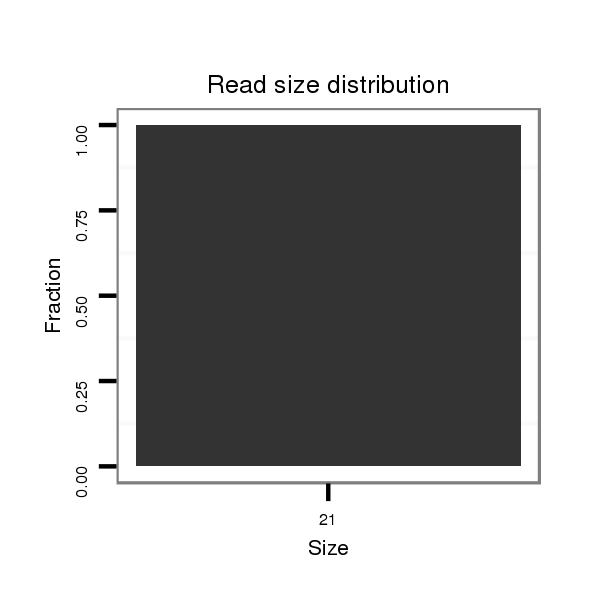
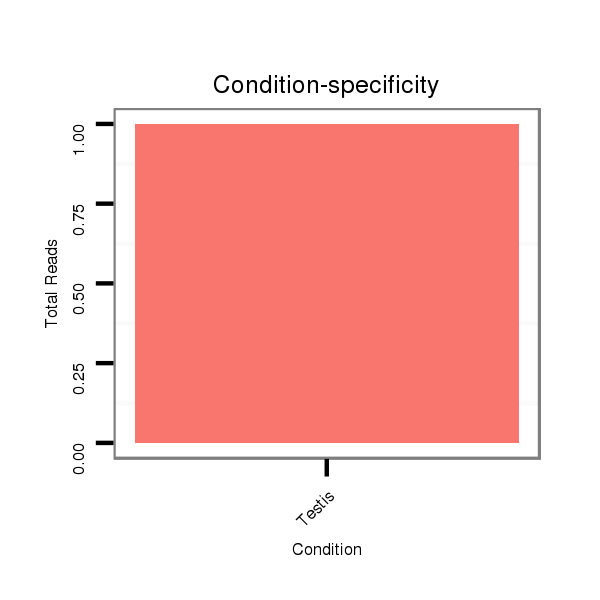
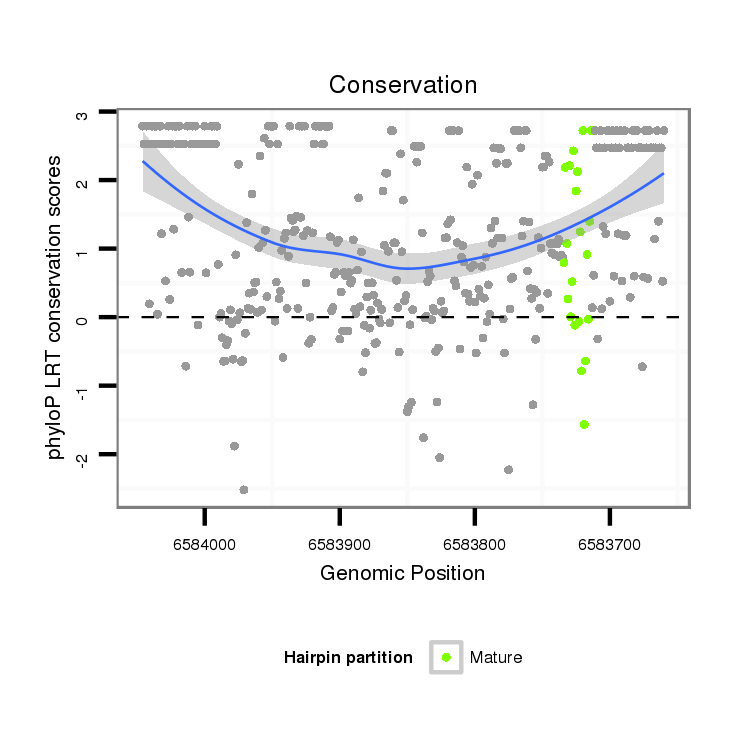
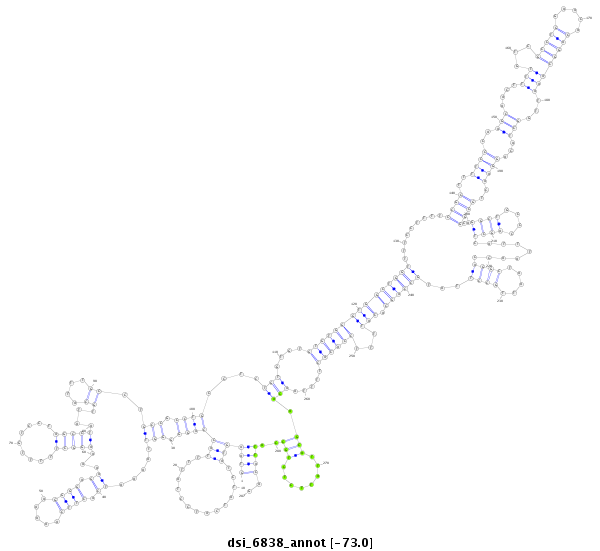
| droSim2 |
3l:6583660-6584046 - |
dsi_6838 |
AGCACCCAGTATGTGCGCCACGTCAACTCACCATACCACACCCCCGAGCCGTAAGTATCT--ATC-AT------CCTA----------------------------T--------------------------TTTACC---AGGG-----GG------TTAAAATCAC-----TTCAAAAAGGGG---AGAA-AAAGG-------CTTTTTGTCCTA-----------------------GCCTAATTGCATTTGCCTGC---CG----------------CCTGCCTCTGTGTGTG--TGTGGGTGG--------CTGGTTT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CCAAGCAACTTTTG--TCGCC-TG------------------GAAC-----------ACAGGCAAATTGGCT------A------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CC--AG---CTCATTTAACC-----------AGGCTAATTGCCTTATGC-----CAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ATTATT----------------CTCT-GCC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droSec2 |
scaffold_2:6645272-6645663 - |
|
AGCACCCAGTATGTGCGCCATGTCAACTCACCATACCACACCCCCGAGCCGTAAGTATCT--ATC-AT------CCTA----------------------------T--------------------------TTTACT---AG-G-----GG------TTAAAATCAC-----TTCAAAAAGGGG---AGAA-AAAGG-------CTCTTTGTCCTA-----------------------GCCTAATTGCATTTGCCTGC---CT--------GCCTGCCGCCTGCCTCTG--TGTG--TGTGGGTGG--------CTGGGTT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CCAAGCAACTTTTG--TCGCC-AG------------------GAGC-----------ACAGGCAAATTGGCT------A------------------------CAGGAACTGCGG-----CAG-------------G----CCGT-CC--AG---CTCATTTAACC-----------AGGCTAATTGCCTTATGC-----CAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ATTATT----------------CTCT-GCC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| dm3 |
chr3L:6708885-6709279 - |
|
AGCACCCAGTATGTGCGCCACGTTAACTCACCATACCACACCCCCGAGCCGTAAGTATCT--ATC-AT------CATA----------------------------T--------------------------TTTACT---AGGG-----GG------TTAAAATCAC-----TTCAAAAAGGGG---AGAA-AAAGG-------CTCTTTGTCCTA-----------------------GCCTAATTGCATTTGCCTGC---CT--------GCCTGCCGCCTGCCTCTGTGTGTG--TGTGGGTGG--------TTGGGTT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CCAAGCAACTTTTG--TCGCC-TG------------------GAAC-----------ACAGGCAAATTGGCT------A------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CC--AG---CTCATTTAACC-----------AGACTAATTGCCTTATGC-----CAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ATTATT----------------CTAT-GCC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droEre2 |
scaffold_4784:4655029-4655432 + |
der_720 |
AGCACCCAGTATGTGCGCCACGTCAACTCACCATACCACACCCCCGAGCCGTAAGTATCT--GAC-CT------CCTA----------------------------T--------------------------TTTACT---GGGG-----GG------CTAAAATCAC-----TTCAAAG-G-GG---AGAA-AAAGG-------CTCTTTGTCGCA-----------------------ACCTAATTGCATTTGCCTGC---CG----------------CCTGCCT--GT----G--TGTGGGTGG--------CTGGGTT-----------------------TCCTGTTCCGC-------------------TTTTCCG---CTTTTCCGCC---------TTTTC-------CCGAGCAACTTTTG--TCGCC-TG------------------TGTT-----------CCAGGCAAATTGGCT------G------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CC--AG---CTCATTTAACC-----------AGGCTAATTGCCTTATGC-----CAGCACATTTTCCCATTTTCCACAC--TTTT-AACAAG---------------C-ATTATT----------------CTCT-GCC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droYak3 |
3L:7293187-7293589 - |
|
AGCACCCAGTATGTGCGCCACGTCAACTCACCATACCACACCCCCGAGCCGTAAGTATCT--ATC-AT------CCTG----------------------------T--------------------------TTCACG---GGGG--C--GG------TTAAAATCAC-----TTCAAAA-GGGG---AGAA-AAAGG-------CTCTTTGTCCTA-----------------------GCCTAATTGCATTTGCCTGC---CT--------GCCTGCCGCTTGCTT--GTGTGTGAAAGTGGGTAG--------CTGGGTT-----------------------TCCTTTTCCGC-------------------TTTTCCA---CTT------------------TGC-------CCGAGCAACTTTTG--TCGCC-TG------------------GCAC-----------ACAGGCAAATTGGCT------G------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CC--AG---CTCATTTAACC-----------AGGCTAATTGCCTTATGC-----TAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ATTATT----------------CTCT-GCC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droEug1 |
scf7180000409711:4273084-4273489 - |
|
AGCACCCAGTATGTGCGCCACGTCAACTCGCCCTATCACACCCCCGAGCCGTAAGTATCT--ATC-AT------CCAC--------TCT--T----TATATATACAT--------------------------TTTCCTAG-AGGG-----GG------TTAAAATCAC-----TTCAAAA-GGGG---GG--AAAAGT-------CTCTTTGTCCTC-----------------------ACCTAATTGCATTTGCCTGC---CT-----------------CTGCCTCTG--TGTG--GGCGGGTAG--------TGGTGTT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CGAGGCAACTTTTT--T-GCC-TG------------------GTAC----------GCCAGGCAAATTGGCT------A------------------------CAGGAACTGCGC-----CAG-------------G----ATGG-CC--AG---CTCATTTAACC-----------CGGCTAATTGCTTTATGT-----TAACA--------CATTTTCCACACACTTTT-AACAAG---------------C-ATTATTA-------TTATT---CTCT-CTC--GTTATAGTGACTCCATTCACGAGCTGCTGGGTCACATGCCGCTGCTGGCCGATCCCA |
| droBia1 |
scf7180000302193:4267154-4267540 - |
|
AGCACCCAGTACGTGCGCCACGTCAACTCGCCCTACCACACCCCCGAGCCGTAAGTATCT--ATT-AT------CT---------------A----TATATATACAT--------------------------TTTGCC---AGGG-----GG------TTAAAATCAC-----TTCAAAA-GGGG---AG--AAAAGG-------CTCTTTGTCCTA-----------------------GCCTAATTGCATTTGC-TGC---CT--------GC---------------------G------TGTGC--------CGGGGCT-----------------------TCCTTTTCCGC-------------------CT------------------------------G----C---CGAGCCAACTTTCTG-T-GCCTCG------------------GAAC-----------ACAGGCAAATTGGCT------A------------------------CAGGAACTGCGCTGGAACAG-------------G----CTGC-CC--AG---CTCATTTAACC-----------AGGCTAATTGCTTTATGC-----CAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-GTTTTTCCCCG----------TCATT-ACC--ATTACAGCGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droTak1 |
scf7180000415809:656324-656702 - |
|
AGCACCCAGTATGTGCGCCACGTCAACTCGCCCTACCACACCCCCGAGCCGTAAGTATCT--AAC-AT------CT---------------A----TATATATACAT--------------------------TTTACC---AGGG-----GG------TTAAAATCACTTCACTTCAAAAAGGGG---GGAA-AAAGGACTCTTTCTCTTTGTCCTA-----------------------ACCTAATTGCATTTGCCTGC---CATT-----------------------GTGTGCG------AGTGC--------CGGTGCT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CTAACCAACTTTTG--CTGC--------------------------------------CAGGCAAATTGGCT------A------------------------CAGGAAC-------------------------------------C--AG---CTCATTTAACC-----------GGGCTAATTGCTTTATGC-----CAACA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ATTATTT----------TCC-CC-TT-CTC--ATTACAGTGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droEle1 |
scf7180000491193:1557734-1558126 - |
|
AGCACCCAGTATGTGCGCCACGTCAACTCGCCCTACCACACCCCCGAGCCGTAAGTACCC--CTC-AT------CTTA----------T--ATA--TAT--ATATAT--------ATGAACAC------TCTTTT-CG--GAGGGG-----GG------TTAAAATCAC-----TTCAAAA-GGGG-------G-AAAG-------CTCTTTGTCCCA-----------------------GCCTAATTGCATTTGCCTGC---CT--------TGCT-----------------------------------------GGC---------ATCA----------TTTCCTTTTCCGC-------------------CTTTT---------------------------GC-------GTGGAAAACTTTTC-----CC-TG------------------GAAC-----------TCAGGCAAATTGGCT------G------------------------CAGGAACTGCGG-----CGT-------------G----TTGG-CC--AG---CTCATTTAGCC-----------AGGCTAATTGCTTTATGCG----GAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-GTTC---------GTTCTC---CTCT-GCCATATTACAGCGATTCCATTCACGAGCTGCTGGGTCACATGCCGCTGCTGGCCGATCCCA |
| droRho1 |
scf7180000779207:32854-33249 - |
|
AGCACCCAGTACGTGCGCCACGTCAACTCGCCCTACCACACCCCCGAACCGTAAGTACCC--ATC-GC------CTTC------------------TATATATACAT----------------------TCGTTT-------GGGG-----GG------TAACAATCAC-----TTCAAAA-GGGG-------G-AAAG-------CTCTTTGTCCTA-----------------------ACCTAATTGCATTTGCCTGC---CT-------------------------GTGTGTG--AATGGGTGG--------CGGCGTT-----------------------TCCTTTTCCGC-------------------TTTTCCG---CTT--C---------------TGCGAGGAAGCGAGGAAACTTTT------CC-TG------------------GAGC-----------CCAGGCAAATTGGCT------G------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CC--AG---CTCATTTACCC-----------AGGCTAATTGCTTTATGCG----GAGCA--------CATTTTCCACAC--TTTT-AACAAG---------------C-ACTATT----------------CTCT-GCC--ATTACAGCGATTCCATTCACGAGCTGCTGGGTCACATGCCGCTGCTGGCCGATCCCA |
| droFic1 |
scf7180000453929:888523-888912 + |
|
AGCACCCAGTACGTGCGCCACGTCAACTCGCCCTACCACACTCCCGAGCCGTAAGTATCC--ATC-AT------CTGA----------T--ATAT-TAT--ATATAA--------------------------CTTCTGAAAGGGG-----GG------TTAAAATCAC-----TTCAAAG-GGGG---GGAGA-AAAG-------CTCTTTGTCCCA-----------------------GCCTAATTGCATTTGCGAA------------------------------------------CGGGTGGCTGCCGGGCTGGGCT-----------------------TCCTTTTCCGC-------------------TTTTC------------------------------------CGGGGCAACTTTTT--C--CC-CA------------------GAGC-----------CAAAGCAATTTGCCT------G------------------------CAGGAACTGCGG-----CAG-------------G----CTGC-CTGGGG---CTCATTTAACC-----------TTGCTAATTGCTTTATGCG----GAGCA--------CATTTTCCACAC--TTCT-AACAAG---------------C-ATTATT----------------TTGT-GTC--ATTACAGCGATTCCATTCACGAGCTTCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| droKik1 |
scf7180000302510:427909-428255 - |
|
AGCACCCAGTATGTGCGTCACGTCAACTCGCCGTATCACACCCCCGAGCCGTAAGTAACT--TT---T------CT---------------A----TATATATACAT--------------------------TTTCC-------------GG------TT----CCAC-----TTCAAAG-GGGCGAAAG--A-AAAG--TG---TCCTTTGTTCGTG---AA-----------------GCCTAATTGCATTTGCCTGC---CT-----------------------------------------------------CGGTT-----------------------TCCTCTTCCGC-------------------CTTTTC--------------------------G----C---CGGGCTAACTTTTA--C-----------------------------------------CCGGCAAATTGGCTGCT---G------------------------CAGGAGC---------------------------------------------CTCATTTGTCC----------CCTGATAATTGCTTTATGC-----TCTGG--------CATTTTCCACAC--TTTT-AACAAG---------------CCATTATTA----------TTG-TG-AT-TTA--CTTACAGTGACTCCATCCACGAGCTGCTTGGCCACATGCCCCTGCTGGCCGATCCCA |
| droAna3 |
scaffold_13337:20204712-20204856 + |
|
AGCACTCAGTACGTCCGCCACGTCAACTCGCCGTACCACACCCCCGAGCCGTAAGTACTT--TAT-TTCATTCTCT-------------AT-------------------------------------------T-GG--ATG--G-----TGTA----AGAAAATCAC-----ATCAAAG-GGGG-------A-AAAG-------CTCTTTGTTCGC-----------------------CCCTAATTGCATTTGCCTAC---C----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396529:208443-208868 - |
|
AGCACTCAGTATGTGCGCCACGTCAACTCGCCGTACCACACCCCCGAGCCGTAAGTACTT--TAT-TTTATTCTCT-------------AT-------------------------------------------T-G---GATGGTGATGGTGTAA----GAAAATCAC-----ATCAAAG-GGGG-------A-AAAG-------CTCTTTGTTCGC-----------------------CCCTAATTGCATTTGCCTAC---CC-T-----------------------GA----G--AGATGATGG--------T-TGGTTTCCTGCCACTA----------CCACCTCTTCCTC-------------------C-------------------GGTGGCGCTT-------C---CGAGCTAACTTTT------CC-TG------------------GA-----------------GCCAATTTGCT------AACG-C----CAACGGCAGTGGGAGCAGGAACTGCGG-----CCT-------------T----CTGCCCC--AG---CTCATTTGCCC----------CGGGCTAATTGCTTTATGT-----TCGCA--------TATTTTCCACAC--TTTT-AACAAG---------------C-ATTATTTCATTCTTTTTT-GTTTTTTTCCC--ATTACAGTGACTCCATTCACGAGCTGCTGGGTCACATGCCCCTGCTGGCCGATCCCA |
| dp5 |
XR_group8:5222447-5222849 + |
|
AGCACCCAGTATGTGCGCCACGTCAACTCGCCCTACCACACCCCCGAGCCGTAAGTGTCG--CCC-GC------------------------------------------------------------------------------CACCCCGCAACA--TGAAAG--C-----ATCAAAG-G-GG-------A-AAAG-------CTCTTTGTGCGC-----------------------CCCTAATTGCATTTCGTTGG---CACT-----------------------------G--AATGGGCGG---------TGGGTTTCCTG----------------------CTTCCTG-------------------CTT-C--CTGC--------------------TG----GCGG-CTAGCAACTTTTC--TCTTG-TT------------------CGAG-----------AGAAGCAAATTGGCTTCAGCTG------------------------CGGGAAG--CAG-----CAGCAGCAGCAG----G----ATAC-CC--ATCTTCTCATTTTGCC-----------TGGCTAATTGCTTTATGTGCTGCGCATA--------TATTTTTCACAC--ATTT-TCTAAC---------------C-ATTATTTCTCGC--TTTT-CCTCTTTCCTC--ATTGCAGGGACTCCATCCACGAGCTGTTGGGACACATGCCGCTGTTGGCCGATCCCA |
| droPer2 |
scaffold_38:469584-469979 - |
|
AGCACCCAGTATGTGCGCCACGTCAACTCTCCGTACCACACCCCCGAGCCGTAAGTGTCG--CCC-GC------------------------------------------------------------------------------CACCCCGCAACA--TGAAAG--C-----ATCAAAG-G-GG-------A-AAAG-------CTCTTTGTGCGC-----------------------CCCTAATTGCATTTCGTTGG---CACT-----------------------------G--AATGGGCGG---------TGGGTTTCCTG----------------------CTTCCTG-------------------T------------------------------TG----GCGG-CTAGCAACTTTTC--TCTTG-TT------------------CGAG-----------AGAAGCAAATTGGCTTCA---G--CGGGAAGCAG------------CAGCAGCAGCAG-----CAG-------------G----ATAC-CC--ATCTTCTCATTTAGCC-----------TGGCTAATTGCTTTATGTGCTGCGCATA--------TATTTTTCACAC--ATTT-TCTAAC---------------C-ATTATTTCTCGC--CCTT-CCCCTTTCCTC--ATTGCAGGGACTCCATCCACGAGCTGTTGGGACACATGCCGCTGCTGGCCGATCCCA |
| droWil2 |
scf2_1100000004729:490262-490639 + |
dwi_1908 |
AGCACACAGTATGTGCGTCATGTCAACTCGCCTTACCACACTCCCGAACCGTAAGTGTTGAAACA-AA------AAAC--------AAA--AAAC-CAA--AA----------------------------------------------------------ACAAAAAT-----TTCAAAA-GTAG-------A-AAAG-------CTTTTTGTGCCCCATGAAGCTAGCTTCACTCGCTTCCCTAATTGCATTTGAA-GCTGCAAAAAGGAAA--GAG--------------ATGCG------GATGC--------TG--------------------------------GTTGGGC-------------------CTT-------------------------------------------------------------GATGTAGTTAGAATGGGGGGAGAACAAAGTTCAAGCAGGAAAATTGCTT--------------------------------------------------------------------------TG--AG---CTCATTT-GCC-----------ACGTTAATTGCTTTATATGC---TAACA-------------------------------ATGGACCGATTTTTTTTC-CCTC---------CTTTTC---CTCT-AT---TTTGCAGCGATTCTATTCATGAACTGTTGGGACACATGCCTTTGCTGGCCGACCCCA |
| droVir3 |
scaffold_13049:1043424-1043826 - |
dvi_17766 |
AGCACACAGTATGTGCGTCACGTCAACTCGCCCTACCACACGCCCGAGCCGTAAGTATAT--ACT-AT------ATAC--------AAT--ATAC-TAT--ATATAT--------------------------CTGCGTAGAG-C----------ATATTTA----AAC-----TTCAAAG-GC-----------AGAG-------CTTTTTGTGCGTC----------------------CTCTAATTGCATTTGCCTGC---CATT--------TTG--------------GTGCG------GCTGC------------------TC----------CAACAGCTTCCGCTTCCGTCGGCAGCTTCTCTTTCTCTTTCTCTG-TGC--TTCCGCC-----------TG----CC--AGTGGCAACTTTTA--TTGTG--------------------------------------CGCCAAATTAGTT------T------------------------AACAAA--GCAG-----CAGCACTTTTAGACACGCCCACAGC-CC--AG---CT-----------ATTATTTAACGCCTAATTGTTTTGCC-------------------TCTTTTTC---------------------------------------T----------------TTCT-CTC--CTTGCAGCGATACCATCCACGAGCTGTTGGGTCATATGCCTTTGTTGGCCGATCCAA |
| droMoj3 |
scaffold_6680:237520-237941 + |
|
AGCACCCAGTATGTGCGTCATGTCAACTCGCCCTTCCACACCCCCGAGCCGTAAGTATAT--ATA-AC------TATGTCTATGTAAAT--ATACATAT--ATATATATGTATATATGTATACATACATATATA-------TG-T----------ATATTTG----AAC-----TTCAAAC-G----------C-AAAG-------CTCTTTGTGCGCG----------------------CTCTAATTGCATTTGGCTGC---CATT--------TTG--------------AGGCG------GCAGC--------C-GGC---------AGCAGCAGCAGCAGCTTCCGCTTCCGCCAGCT---G----------CTC-C--TTGC--TT--------------------------CCGACCAACTTTTA--TTGTG-T-------------------------------------CGCAAATTAGTT------T------------------------AACA--ATGTGC-----CAG-------------G----CAG--CC--AG---CACTTTAAGCCCCATTATATAATGGCTAATTGTTTTCT------------------------TTCCTTTC--TCT--------------------------------TTC---GTTCTCCTTCTCT-CC---CTTTTAGCGATACCATTCACGAGCTGTTGGGTCACATGCCGCTGCTGGCCGATCCCA |
| droGri2 |
scaffold_15110:2199762-2200167 - |
|
AGCACCCAGTATGTGCGTCACGTCAACTCGCCGTACCACACACCCGAGCCGTAAGTAGTC--CACCCCCATTATCT-------------AC------------TCGT--------------------------TTT--------------------------GCATAAC-----ATCAAAG-G-CG-----AAA-AAAG-------CTTTCTCTGC-C-----------------------CTCTAATTGCATTTGCCAGC---CATT--------T----------------GTGCT------GTGGC------------------TC------GAAGCAGCAGCTTCCGTTTCCGTCTGCT---TCTGCTTCTG-----------C--TTCT---GCTTCTGCTTCTG----CAGTCTGGCCAACTTTTTGATTG--------------------------T-----------GCTGGCAAATTAGTTT-A---G--C--------------------------------C-----CAGCAC---------------TTGC-TG--AG---CT-----------ATTATTTAGGGGCTAATTGTTTTATGT-----GCACA--------CA-----CACAT--TTTGTAACACG------TTTCCTTTGC-CTTT---------GTTCTGC-TCTCT-T-C--CCTTCAGCGATACCATTCATGAGCTCTTGGGTCATATGCCGCTGCTGGCGGATCCTA |