
ID:dsi_6461 |
Coordinate:2r:5290260-5290409 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
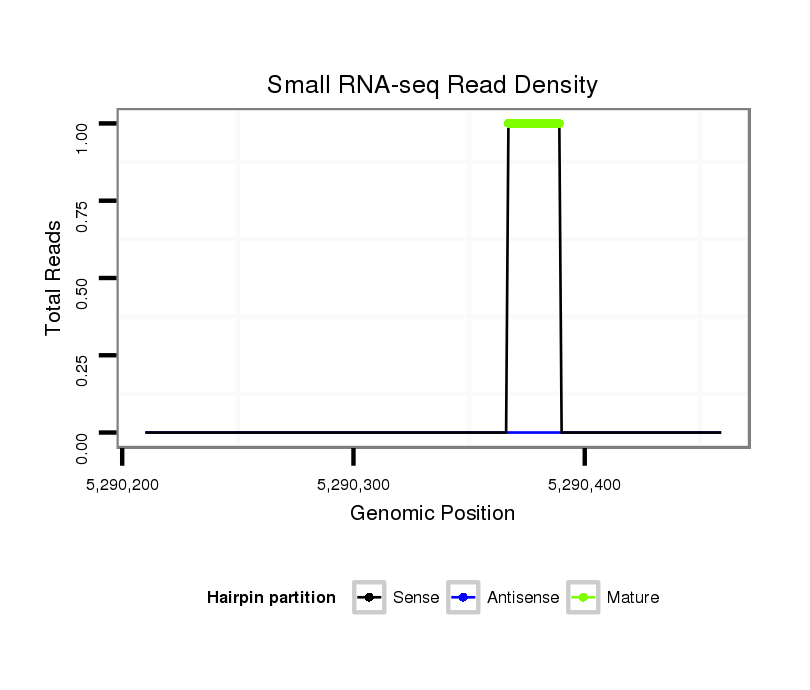
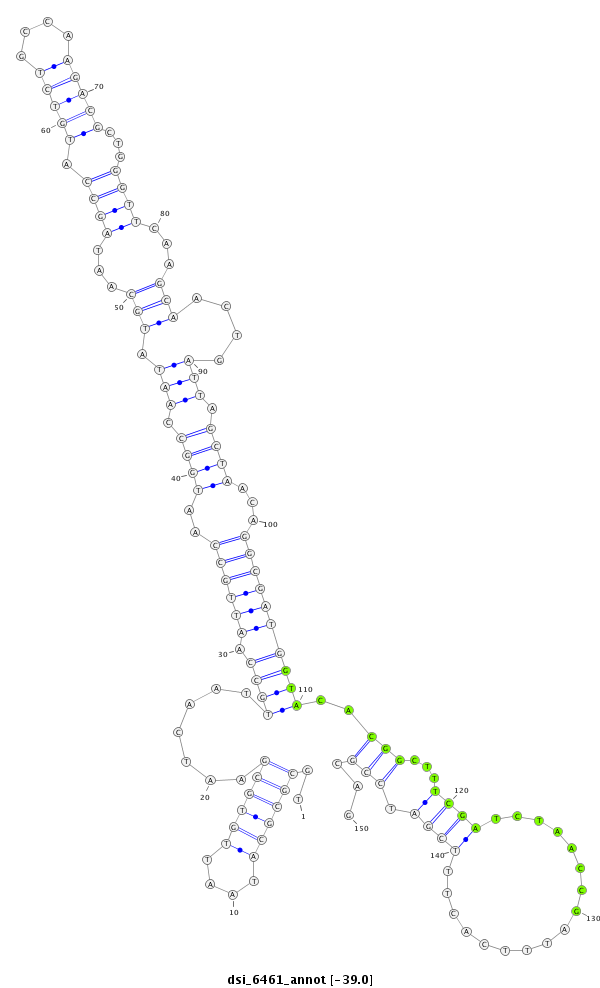
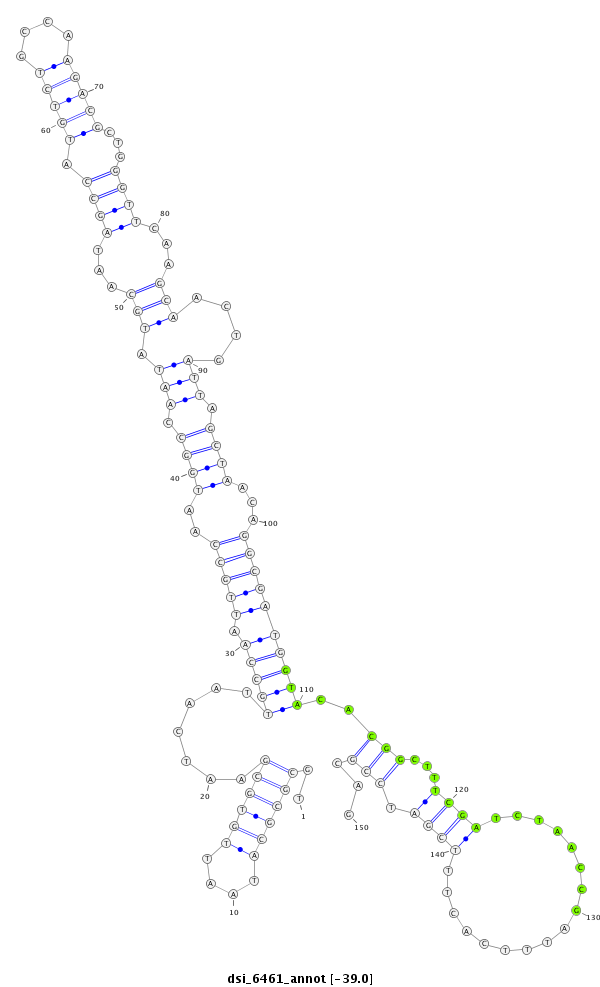
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
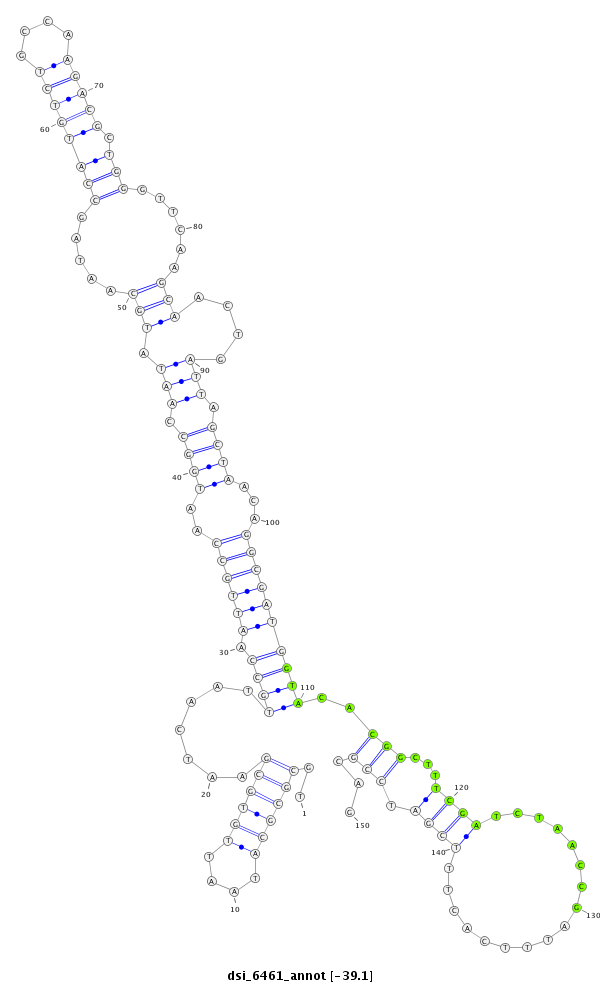
| -39.1 | -39.0 | -39.0 |
 |
 |
 |
CDS [2r_5290410_5290665_+]; exon [2r_5290410_5290791_+]; intron [2r_5289930_5290409_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCGCGGCTTAGGAATTTCGCAGGCGGCGGTTGGCATTTAATTCAGCGTAATGCGCGCATAATTGTGCGAATCAATTGCCAATTGCCAATGGCCAATATGCAATAGCCATGTCTGCCAAGACGCTGGGTTCAAGCAACTGATTAGCTAACAGGCGATGGTACACGGCTTTCGATCTAACCGATTTCACTTTCGATCCGCAGCGAGCACCACACCATCCACGTACCCTACAAGGTGCACACGGTGCACCACC **************************************************..((((((....)))))).......(((((.(((((..((((.(((.(((.....((((((((....))))).)))......)))....))).))))...))))))))))..(((...((((.................)))).)))...************************************************** |
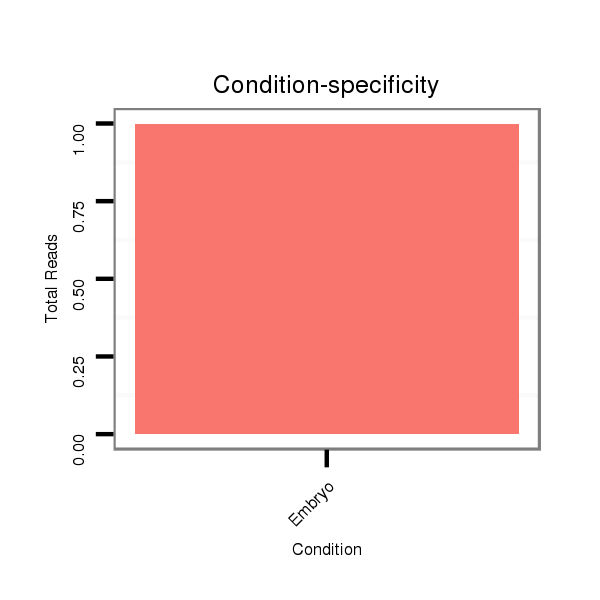
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR902009 testis |
M023 head |
|---|---|---|---|---|---|---|---|---|
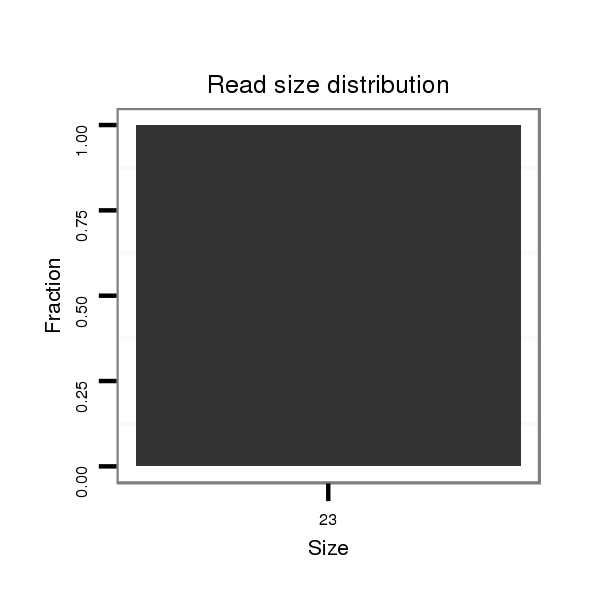
| .............................................................................................................................................................GTACACGGCTTTCGATCTAACCG...................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ..........GGAATTTCGGACGCGGCGGTT........................................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 |
| ..........GGAATTTCGGACGCGGCGTTT........................................................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 |
| ..................GCAGTCGGAGGTTGGGATT..................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
|
AGCGCCGAATCCTTAAAGCGTCCGCCGCCAACCGTAAATTAAGTCGCATTACGCGCGTATTAACACGCTTAGTTAACGGTTAACGGTTACCGGTTATACGTTATCGGTACAGACGGTTCTGCGACCCAAGTTCGTTGACTAATCGATTGTCCGCTACCATGTGCCGAAAGCTAGATTGGCTAAAGTGAAAGCTAGGCGTCGCTCGTGGTGTGGTAGGTGCATGGGATGTTCCACGTGTGCCACGTGGTGG
**************************************************..((((((....)))))).......(((((.(((((..((((.(((.(((.....((((((((....))))).)))......)))....))).))))...))))))))))..(((...((((.................)))).)))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR902009 testis |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................ATTGGCCAAAGGGAAAGC.......................................................... | 18 | 2 | 9 | 0.78 | 7 | 1 | 6 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CCAAGTTCGGTGACTA............................................................................................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................TTAACACGCTTAGTTG............................................................................................................................................................................... | 16 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TTGTGGTGTAGTAGGTGC.............................. | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................TAACACGCTTAGTTGACA............................................................................................................................................................................ | 18 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GGGATGTTTCACATGTGAC......... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TCGATTGGCCGCTAC............................................................................................. | 15 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................ATGTGCCGGGAGCTAG............................................................................ | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................CTAGGGGCATGGGAT....................... | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................CGCTCGGGATGTCGTAGGT................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................CGGAAACCAAGTCGCATT........................................................................................................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................TCCTGTGACCCAGGTTCGT................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................AGTGAAAGCTAGCCG.................................................... | 15 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................CGGTTGCCGGCTATAC....................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/23/2015 at 12:05 PM