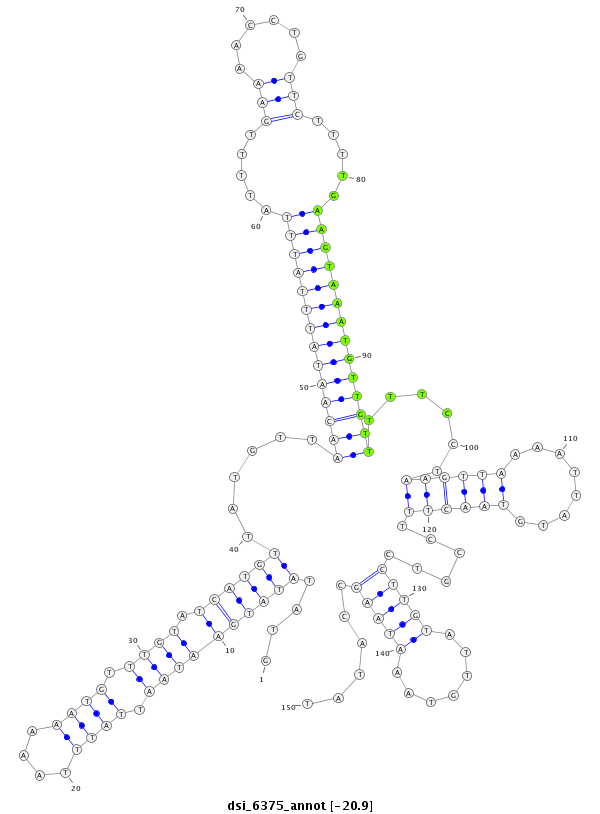
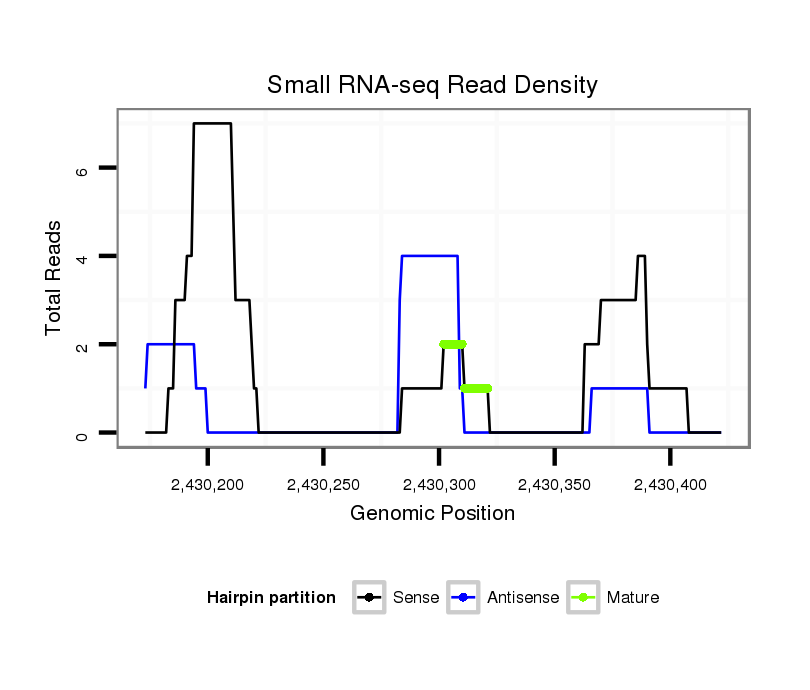
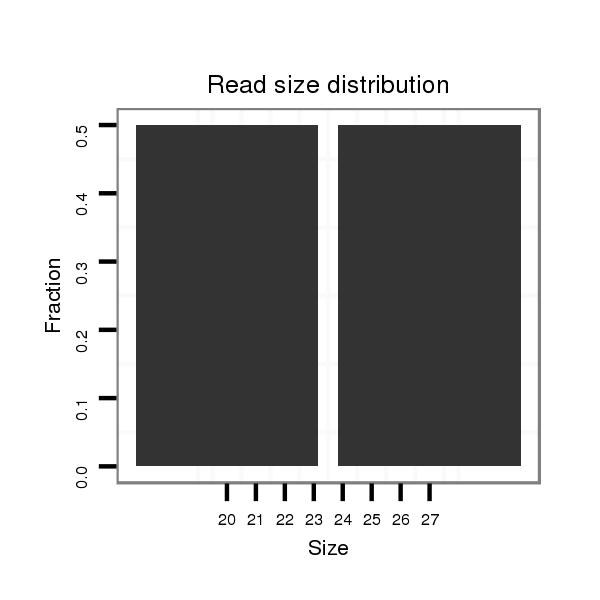
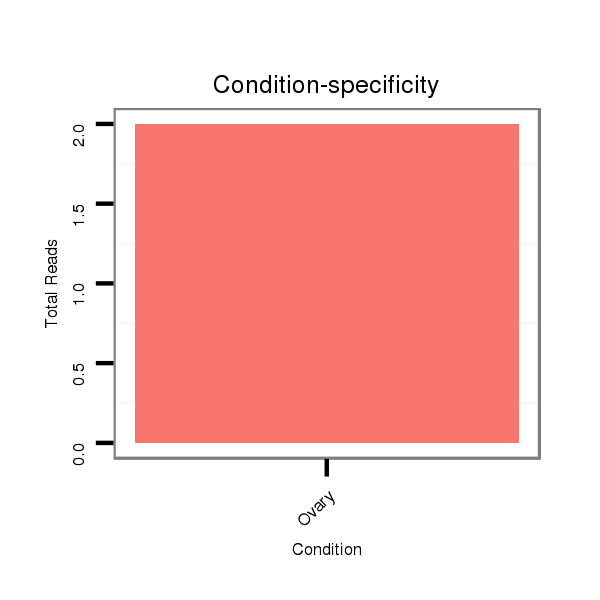
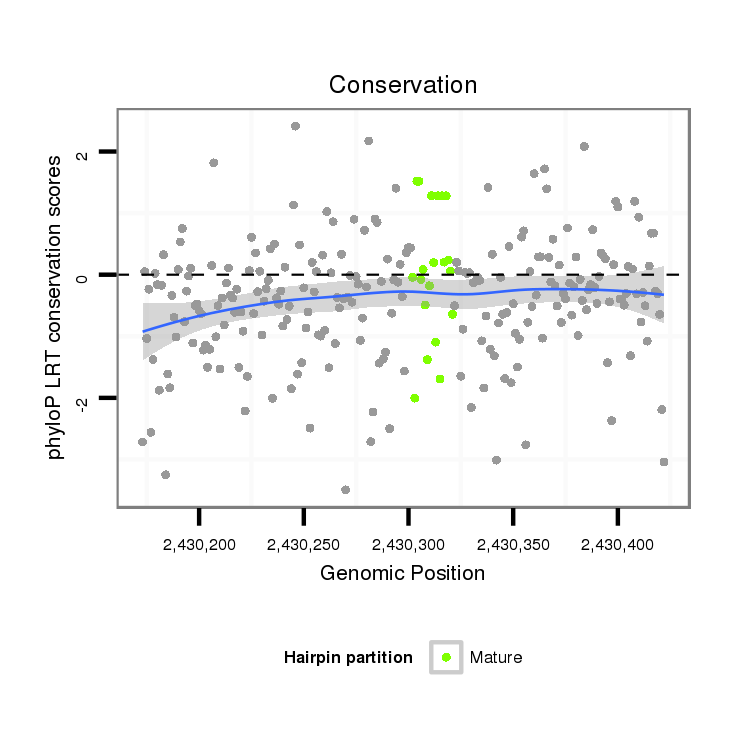
| droSim2 |
2r:2430173-2430422 + |
dsi_6375 |
AAGTCAC------ACAAGATAC-CA--------------------------------TTCTG-----GGAATTGGCTAAGGGA-----T--TAAT---ATCCAGGTATATA--TGA------------------------------------------------ATAATTATTTAAAAA---TGTTTGTATC--A-----TG--------------------------------------------------------------------------TTATGTTAACAATATTTATTTATTTTG-----------------------------------------------A-----------AA--ACCTGTTCTTTTGA-AG--------------------------------------------T-------------------------------------------------------------------------------------------AAATGTTGTTTTTCCTAA------------------------------------------------------------------------------GTTAAAATTATGTA------------------------------AC-----TTTCCGTCCTTG---------------------------------------------------------------------TATTGTAAAT----------------------------------AAGCCATATCCGTTCGTCCGTTCGTCTGTCC--------AGGGACATCGATGACAT--CATAAATAAAC |
| droSec2 |
scaffold_26:60862-61102 + |
|
AAGTCAC------ACAAGATAC-CA--------------------------------TTCTG-----GGAATTGGCTAAGGGA-----T--TAAT---ATCCAGGTATATA--TGA------------------------------------------------ATAATTATTTAAAAA---TGTTTGTATC--A-----TG--------------------------------------------------------------------------TTATGTTAACAATACTTATTTATTTTG-----------------------------------------------A-----------AA--ACATGTTCTTTTGA-AG--------------------------------------------T-------------------------------------------------------------------------------------------AAATGTTGTTTTTCCTAA------------------------------------------------------------------------------GTTATAATTATATA------------------------------AC-----GCTCCGTCCTTG---------------------------------------------------------------------TAAGGC-------------------------------------------ATATCCGTTCGTCCGTTCCTCTGTCC--------AGTGACATCGATGACAT--CATAAATAAAC |
| dm3 |
chr2R:1416269-1416457 + |
|
AAGTCAC------ACAAGATAC-CA--------------------------------TTCTG-----GGAATTGGCTAAGGGA-----T--TAAT---ATCCAGGTATATA--TGAAAACAAAA-------------------------------------------ATTATTTAAAAAC--TGTTTGTATA--A-----TG--------------------------------------------------------------------------TAATGTTACCAATATTTATTTAGTTTG-----------------------------------------------G-----------AA--ATTTGTTTTTTTGA-AG--------------------------------------------T-------------------------------------------------------------------------------------------ATATGTTATTTTTCCTAA------------------------------------------------------------------------------GTTATAACTATATA------------------------------T----------TGTCCCCG---------------------------------------------------------------------TATT----------------------------------------------------------------------------------------------------------GTA |
| droEre2 |
scaffold_4929:20984716-20984855 - |
|
AAGTCAC------ACAAGATAC-CA--------------------------------TTCTG-----GGAGTTAGCTAAGGGA-----T--TAAT---ATCCAGGTATATA--TGAAATAAGAA-------------------------------------------ATTATTTAAAATT--TGTTTGTATC--C-----TA--------------------------------------------------------------------------TAATGTAAAAAGAATTTTTTTCTTTTG-----------------------------------------------G-----------GAC-GTTTGTTTTCTTTA-A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droYak3 |
v2_chr2L_random_001:311158-311414 + |
|
AAGTCTC------ACAAGATAC-CA--------------------------------TTCTG-----GGAGTTGGCTAAGGGA-----T--TAAT---ATCCAGGTATATATATGAAATCAGAT-------------------------------------------ATTTTTTAAAATT--TGTTTCTATC--A-----TA---------TTA-------------------------------------------------------TA-----TAATGCTAATAATATTCATTTCTTTTG-----------------------------------------------G-----------GT--CTTTGTTCTTGTCA-AG--------------------------------------------T-------------------------------------------------------------------------------------------AAATTGTATTTATCCTAC------------------------------------------------------------------------------GTTG--------TA------------------------------AC-----GCTCAGTACCCG---------------------------------------------------------------------TAGAGAACAAAATTTTTTTATCAGGACTA---------------------CTACCGTTCATTCATTCGTGCATCC--------GATGACATCAAGGACA-------------T |
| droEug1 |
scf7180000409759:423829-424094 - |
|
TATTTTGT-----ATACTGCA--CA--------------------------------GTCCG-----GTTATAATTAAAAAGC-----T--TAAC---AGTCAAA-ATATA------------------------------------------------------TTTAAACTTTCAAAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAACATCTCAGCATTACAATCACTACACGATATTTACCTATTCTATCAA-------------------------------------------------------------------------------------------------------------------------------------------GAATCTGTTGTGTATCATTTCACGTTCGAA-------CTTGT-----TC--A----TAGGCG-----GAATACGAACGTAAGCAT-----------------------CCAACG---------------------------------TAAGCAAGTTGATTGAGCCATGTTCGTCTGTCCGTCCGTCCGTCCGTCCTTAT--GAACGCTGA-GAT--CTCAGGAACTATA |
| droBia1 |
scf7180000302055:18343-18605 + |
|
TAGGAAT------TTTAGGT--------------------------------------------------------------T-----T--AAAT---ATTAAAGTAGACT--TGG--------------------------------------AGAATAT---ATAATTATTTACAAAATATACCACTGTT--G-----TA---------TAATT------------------------------------------------------------TAATGATGACAAATTA---------------------ATGTCTC-TTTTTATCCCTTTTTATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTT-----GC--A----GAGGGTGTAATGATTTCAGTCAGAAGTTTGCAACGCAGTGAAGGAGACGTTTCCGACCCCATAAAGTATATATATTGTTGATCAGCGCCACTAGACGAGTCGATCTAGCAATGTCCGTCTGTCCGTCCGTCTGTC--------------------------------------- |
| droTak1 |
scf7180000415376:31973-32232 + |
|
CTGTTAT------ACATAAAAA-TC---AACAAATAAC-AGTTATTTTTGACTTTTATTCTA-----AAAATCAAATAAACAA-----A--AATCA-TTAC-----------------------------------------------------GGATTTGTAAACTA--------------------CAA--CGGCTTACAACA-----ATAATGGTG-----------------------TATTTCAAAAAATTTTTGGAC--------------------------------------------------------------------------------------------------------------TCTTCTA-AG--------------------------------------------T-------------------------------------------------------------------------------------------GCATGATTTTTTTGGTAAGAAAAATTTACAATAACAGTTATTTTCGGTCCCTGGATTCGAAACAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTTTACTGTGTGATTCGAATGATT--------CGAAACATCTAAGACAG--TGTA----AAT |
| droEle1 |
scf7180000490512:896-1136 + |
|
GGTCA-T------GCTAAGTAC-AA--------------------------------TTATG-----A------------------------------------------------AATAAAAATTT-----TTAA--------TCTGTCCTTG-------TGAACTG----------TT--TGTTTTTATT--A-----TT-------------------------------------------------------------------CG-----TTGTTTTAATTTTATTCTTCTTTT----------------------------------TTATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTT-----GC--A----GAGGGTATTATAATTTCAGTCAGAAGTTTGCAACGCAGTGAAGGAGACGTTTCCGACCCTATAAAGTATA----TTCTTGATCAGCATCACTAGTAGAGTCGATCTAGCCATGTCCGTCTGTCCGTCCGTCTGTCC-------------------------------------- |
| droRho1 |
scf7180000770855:556-856 + |
|
AAATTTTTTCAACAAGATATGC-CA--------------------------------TTCCA-----ACATTCTACCA----------C--TATT---TCCCAGGGTT----------------------------------------------------TTGATTAAAAACCGATAAA---TC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GAGGTGATTTTCTCCGGACTTGGGCCTTGTCATACGGGCGTTTTCAAAAAAATGTTTCAA---------------ACGATA----------------------------------------------------------------------------------------------------------------------------------------------TAT--------TT------------------------------AT-----ACCCGTTACTCGTAGAGTAAAAGGGTATATTGTATTCGTGCAAAAGTATGTAACAGCGAGAAGGAAGCGTTTCCGACCCTATAAAGTATAT---------------------------------ATAGCCATGTCCGTCTGTCTGTCTGTCTGTCC----GTAT--GAACGCTGA-GAT--CTCGGAAACTATA |
| droFic1 |
scf7180000454077:92172-92365 + |
|
AAATGTT------AAAATACA---A--------------------------------TTTAG------TCTAC------TTCT-----T--CATG---AGAACCCTGAAAT--GCT------------------------------------------------CTGA--ATGGCTAAT---A--------C--T-----TT---GTATA-------------------------------------------------------------------------------------------------TATAAAAA--CTC-ATTTGATTTATTTTTGCACACT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T--------------------------------GTCAACAGGAAAGAACTCTTATTTCAAAATAAATAAGGCATTTTAATTTATAATAAAATAAA-----------------------------A-----ACTTCGTTTTTA---------------------------------------------------------------------TGTTCTAA------------------------------------------------------------------------------------------------------ATA |
| droKik1 |
scf7180000302414:478142-478321 - |
|
AAATAAT------ATAATTTAT-ATTTAAACAAGAAAAC--------------------C-----------GCTTGAAAAGTCATTCTT--TTAT---ATTTAAATATATA--AAA------------------------------------------------ATAAATTCTAATAAA---TAAAAATAA--CAACCTATA--------------------------------------------------------------------------ATATATATAAAATATTTATATAATAAG-----------------------------------------------A-----------AA--A-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATATTTTAAATTAATA-------------TTGATTATTTTT--------------------------------------------------------------------------------------------------------------------------------------AATATTAATTTA--------------------------------------TAT |
| droAna3 |
scaffold_13099:223073-223282 + |
|
CAAAAAA------GCAAGAGTTCTA--------------------------------TCTTGTGACGGAGATATGCCGAGTGA-----CATACAT---ATTCCGATACGGA--C--------------------------------------------------ATTACTTCTCACAAAA--TGTCCTT-----T-----TG--------------------------------------------------------------------------TCATCATACCAATATT----TACCTTG-----------------------------------------------G-----------CA--CCTTTTCCTTTCAAGAGTCAATGAATAAAATTGCTTTTC--------------------------------------------------------------------------------GATCAAAAATATG--------------------------------------------------------------------------------------------------------------------------AATTACATA---------------AATT------------------TTTTTGTTCAAA---------------------------------------------------------------------TAGTTTATTT----------------------------------AA----------------------------------------------------------------GCC |
| droBip1 |
scf7180000396730:669189-669331 - |
|
TGATCAT------TAAAGTACT-------------------------------------TTG-----GAAAT---------------------AT---ATGCATGTACATA------------------------------------------------------------TTTAAAAT---TATTCATGTT--T-----TA--------------------------------------------------------------------------TTTTTA-AAGGGAAATAGTTTTATTAA-----------------------------------------------A-----------AA--GC-------------------------------------------------------------------------------------------------------------------------------AA--------------------------------------------------------------------------------------------------------TTTCAA--------AACATTGTTTAGTC------------------------------ATCTGATTTTCTTTCTTTT---------------------------------------------------------------------TATTGCAT------------------------------------------------------------------------------------------------------ACA |
| dp5 |
Unknown_group_201:12145-12316 - |
|
AATCCCA------ACAAACAAA-T----------------------------------------------------------A-----T--CAGT---ATTTCTGTAGGTA--AAA------------------------------------------------ATAATGA-----------------------------------------------------------------------TATTTCAATAAAATTTCGAAGTTTTTTAATAATGTAATA--AAGAAAACCCATTTTTTTCA-----------------------------------------------A-----------AA--AT-------------------------------------------------------------------------------------------------------------------------------ACTGATTTTTTGATCGTCTCTAGGTAGTGCTTTTTTGTTTCA------------------------------------------------------------------------------ATTATTATTCAATT------------------------------AT-----T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTC |
| droPer2 |
scaffold_1321:1071-1300 + |
|
GAATTAA--------TAGGTAA-TG--------------------------------TTTTT-----GCAAAAAAATTAGTAA-----A--TAGC---CTTTAAAAACGTA----------------------------------------------------------TCCTGGGAAC---CACAAACT-TTCTTGCTATATTA-----AGAATGAATCAATTAATGCAAAAAATGTAC-------------------------------------------------------TTCATTTA----------------------------AATCGGTAAGTG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------G------------------------------------------------------------------------------GTTAAAAAGATATA------------------------------AC-----GGTTTGACTTTT---------------------------------------------------------------------GAAAG-------------------------TTGTTAAGTCGATTCAGTTGAGCGGCTCTGTACAT--------------------AAACATTGCTTACAATATAAA----AAC |
| droWil2 |
scf2_1100000004558:556183-556502 + |
|
ATTCTGT------ACAATTTAG-AA---------------------------------------------------------------A--TTTT-GTTTCTGAATATAAG--CAACTACAATA---AATTAT---GGAAAATGATTGTCGTGGAG------------------------------------------------------TCAATGAAG-----------------------TATTACAAAGAAGTGTTTAATTGTTTTCG-----TTATGTCATTGATATCCAATTAATTCGAATCTAAAAAAGTTCTC-ATTTAG-TAATTATTATAC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTT-----GC--A--AAAAGGGTATATTAATTTTGGTCAAAAGTGTGCAACGCATAGAAGGAAGCATTTCCGACCATATAAAGTATATATATTCTTGATCAGCACGACGAGACGAGTTCAAATAGCCATGTCCGTCCGTCCGTCCGTCCGTCC-------------------------------------- |
| droVir3 |
scaffold_12963:14577989-14578109 - |
|
TATTATT------ATTTGATTT------------------------------------TTAG------ACATT------TTCA-----T--TTAT---ATGACGCTATATA--TTT------------------------------------------------CTGATTATTACGAAT---T--------C--A-----AT---TTATA------------------------------------------------------------------TTAAA-AACATATATATGTATTTTTTG-----------------------------------------------A-----------AA--ACTTACACTTTTAA-AG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T |
| droMoj3 |
scaffold_6500:3446897-3447075 + |
|
AATTATT------AAAAAACAA-AA----------GATT--------------------C-----------A-------------------------TTTT-----------------------------------------------------AGCGTTATAAATTATAACTGAAAAA---TCTTTTTTTT--T-----AA---------TAATT------------------------------------------------------------TCG-----------------TAATTAA-------AAAAAATTTTTTTTTTCGTTTTTTTTAAGT--TTACATTTTTAAATTTTTACTTTTTTTTATTTTTTTTA-AA--------------------------------------------T-------------------------------------------------------------------------------------------AA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATTTTTTGTTTCGTTTATCA---------------------------------------ATC |
| droGri2 |
scaffold_15081:2397132-2397295 + |
|
GAGTAAT------GTAACAGCT-------------------------------------GTG-----GAATGTTTGGAAATGATTACTA--TTAA---GCTTGAAGA----------------------------------------------------------------TGGCATAT---T--------CGTTTTGCAT-----------AACAAAATTATTAATATAATATATATATTTTATTTATATATATTT---------------------------------------------------------------------------------------------TTGAGTTCTTATGAG-----------------T-----------------------------------------------------------------------------------------------------------------------------------------TTTGCTGTCTTTCTAAA------------------------------------------------------------------------------GATAAACTTTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AT |