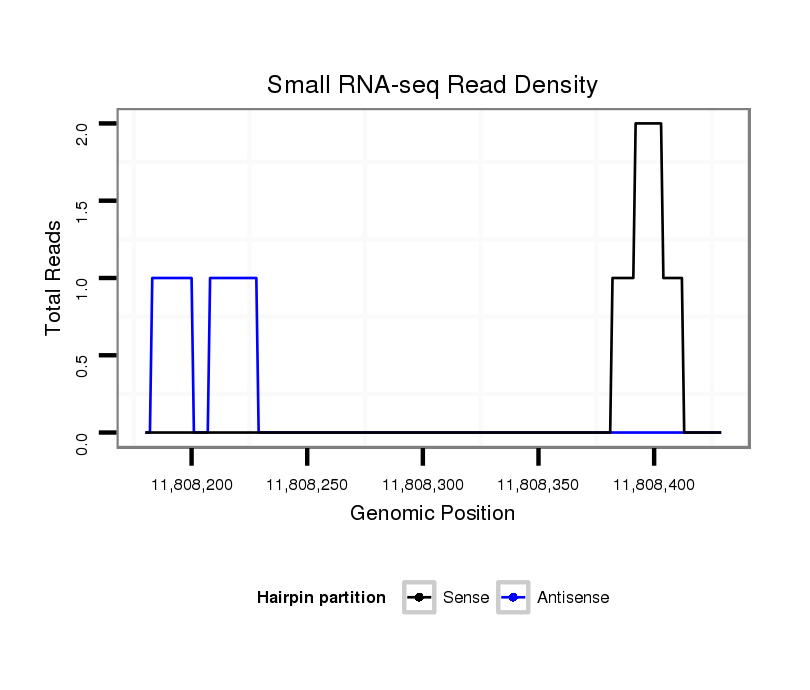
ID:dsi_6292 |
Coordinate:3l:11808230-11808379 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3l_11808380_11808786_+]; exon [3l_11808380_11808786_+]; intron [3l_11808073_11808379_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TGTTTACCTTTTCATAAGGTTTAAATGTACCTTTTCCTTTATCAGAACAGTGTGTTTTCGTTAAAACATGTGATAGAATATCAAATTTGACCTAATTTCTGTATGAAGTCTACTTAGCCTAACCTGGATAACTAGCAAATATACCAAAATTCTATTTAAGCCCTTAGTAACCATGCTTCTAAAACAACTGATTCTTCTAGAATATAACAGCAGAACGTCGTAAGCCAGGGCCGCGCATGAAGAGCTCACT **************************************************.(((((((....)))))))....(((((.((((...(((....(((((..(((((....(((.(((.((......)).))).)))....)))))..)))))......((((.............))))......))).))))..))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................CAGAACGTCGTGAGACAGG..................... | 19 | 2 | 3 | 5.67 | 17 | 0 | 14 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCG................. | 17 | 2 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCC.................. | 16 | 2 | 9 | 2.67 | 24 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................GAACGTCGTAAGCCAGGGCCG................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TATAACAGCAGAACGTCGTAAG.......................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................TTCGTTAAAACATGTAACAGA............................................................................................................................................................................. | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCGCC............... | 19 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCTC................ | 18 | 3 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCGAG............... | 19 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCTCG............... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CAGAACGTCGCAAGACA....................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................AAACATGGGATAGAA............................................................................................................................................................................ | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................CCTGATCTCTGTAAGAAGT............................................................................................................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................GTACGAATTATACTTAGCC................................................................................................................................... | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................GTCGTGAGACAGGGCCGG................ | 18 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CAGACCGTCGTAAGAC........................ | 16 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................TAACCATGCGTCGAAAAAAA............................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................GCAGACCGTCGTAAGACT....................... | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CACTGGATAACTAGGAAAT.............................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ACAAATGGAAAAGTATTCCAAATTTACATGGAAAAGGAAATAGTCTTGTCACACAAAAGCAATTTTGTACACTATCTTATAGTTTAAACTGGATTAAAGACATACTTCAGATGAATCGGATTGGACCTATTGATCGTTTATATGGTTTTAAGATAAATTCGGGAATCATTGGTACGAAGATTTTGTTGACTAAGAAGATCTTATATTGTCGTCTTGCAGCATTCGGTCCCGGCGCGTACTTCTCGAGTGA
**************************************************.(((((((....)))))))....(((((.((((...(((....(((((..(((((....(((.(((.((......)).))).)))....)))))..)))))......((((.............))))......))).))))..))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...AATGGAAAAGTATTCCAA..................................................................................................................................................................................................................................... | 18 | 0 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ............................TGGAAAAGGAAATAGTCTTGT......................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................CAGATGAATCGGATGGGC............................................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................TGGTACGAAGGCTTTGT................................................................ | 17 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................ACCTATTGGTCGTTTA.............................................................................................................. | 16 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................ACGAAGATTTTGTTGG............................................................. | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................GAAACGTTGGTACGAAG....................................................................... | 17 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................TTAGGCCCTATTGATCGTT................................................................................................................ | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................TGGTTTGTATGGTTTTTAGAT................................................................................................ | 21 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................TCCAGCATTCGGCCCC.................... | 16 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:37 AM