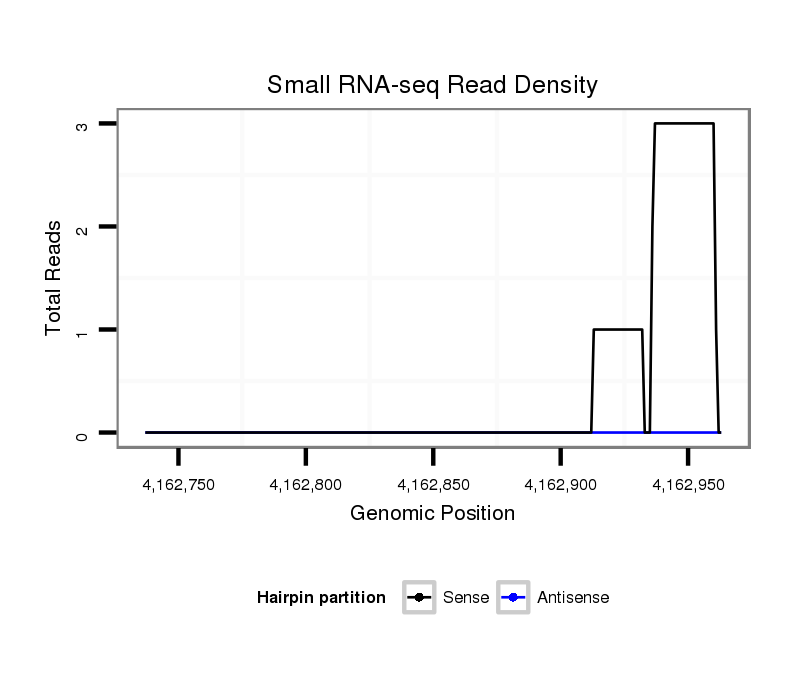
ID:dsi_6033 |
Coordinate:3r:4162787-4162913 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [3r_4162649_4162786_+]; CDS [3r_4162649_4162786_+]; CDS [3r_4162914_4163051_+]; exon [3r_4162914_4163051_+]; intron [3r_4162787_4162913_+]
| Name | Class | Family | Strand |
| (TTCC)n | Simple_repeat | Simple_repeat | + |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------################################################## CCTGTTGAGCTTCCAGGCCTGTCCCGATGTCCAGGGCACCGAGAGTACGGGTAAGTACCGCCTCCTCCGCAGGCGCCAATGACTCATCGCCTTTCTGTGCCCGGCTGGAGCTCAAGACGAACGTCCTTCCTTCCTTCCTTCTTGCTTCCTTTCTTTTCGGTGCTGGTGCTCCTGCAGTCGAGGAGCTGACTTGCCTGGCAACCTGGAAGGACGGCAACTCACGCTAC **************************************************...(((((((.((((.(((..(((((....................))))))))..))))..(((((.(((.............)))..))))).............)))))))..(((....))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR1275483 Male prepupae |
M023 head |
M025 embryo |
SRR902009 testis |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR1275487 Male larvae |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................CTACAGTCGACGATCTGAC..................................... | 19 | 3 | 7 | 3.14 | 22 | 21 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................AACCTGGAAGGACGGCAACTCACGC... | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GTCGAGGAGCTGACTTGCCT............................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................TCGGTGCTGGGGCTCCTGCAG.................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTACGGTCGAGGAGCTGACTTGC................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ACCTGGAAGGACGGCAACTCACGCT.. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GTCGAGGGGCTGACTTGCCC............................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................GTACGGTCGAGGAGCTGACTT................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CTGCAGTCGAGGATCT........................................ | 16 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..............................................ACGGGTGAGTAACGCCT.................................................................................................................................................................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................GGGTAAGAACCGCATCC.................................................................................................................................................................. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........CTTCCAAGGCTGTCCC.......................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................CTTCGTTCCTTCTTG................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GGACAACTCGAAGGTCCGGACAGGGCTACAGGTCCCGTGGCTCTCATGCCCATTCATGGCGGAGGAGGCGTCCGCGGTTACTGAGTAGCGGAAAGACACGGGCCGACCTCGAGTTCTGCTTGCAGGAAGGAAGGAAGGAAGAACGAAGGAAAGAAAAGCCACGACCACGAGGACGTCAGCTCCTCGACTGAACGGACCGTTGGACCTTCCTGCCGTTGAGTGCGATG
**************************************************...(((((((.((((.(((..(((((....................))))))))..))))..(((((.(((.............)))..))))).............)))))))..(((....))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................TGGCGGCGGAGGCGTCCG......................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TCACCCTTCCTGCCGTTGAGT...... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................GAGTTGTGGTTGCAGGAAGGAC............................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................ACCGCTGGACCTTCC................. | 15 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 2 |
| ......................................................................................................................................................................................................GTTGGACCATCATGCCG............ | 17 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................AGGAAAGACAAGCAACGACTA............................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 |
| .........................................................GCCGGAGGAGGCGACCGC........................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AGATTCCTGCCGTTGA........ | 16 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
Generated: 04/23/2015 at 09:28 PM