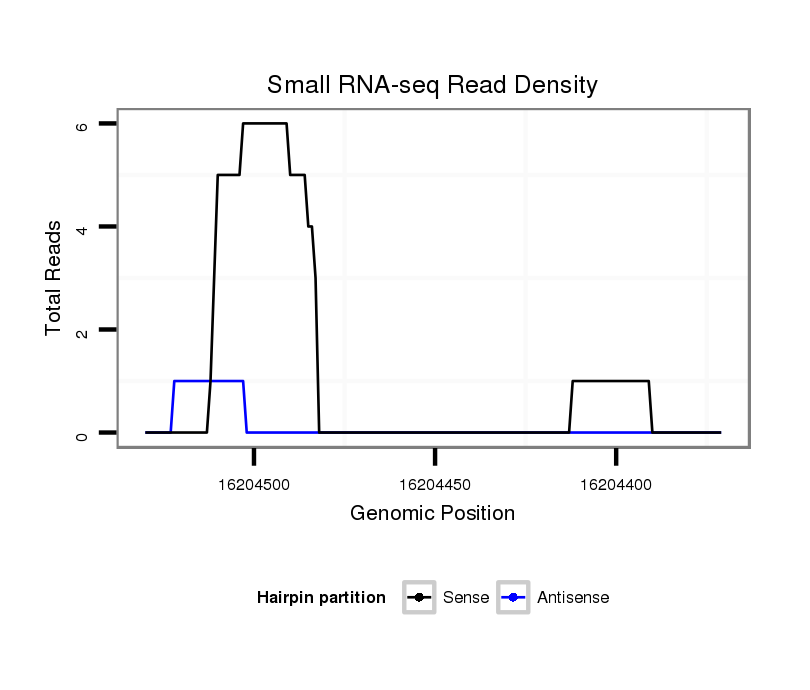
ID:dsi_5986 |
Coordinate:3l:16204421-16204480 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3l_16204329_16204420_-]; exon [3l_16204253_16204420_-]; CDS [3l_16204481_16204887_-]; exon [3l_16204481_16204887_-]; intron [3l_16204421_16204480_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------################################################## CATGGAGGGGTGTCAGACGTTCAAGGATTGTCTGCAAAACACCAGCATTGGTAAATAGTTTCTTTCTCTGGGTTTGTACATTTTTCTGAAGTTTTTACATTTCTCCGCAGGTAAATGGTTTTATGACGTCAAGGCGTTGGTGGAGAAGAATCGTGGGCAG **************************************************.................(((((..((((.(((((...)))))...)))).....)).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR902009 testis |
M024 male body |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................TTCAAGGATTGTCTGCAAAACACCAGCAT................................................................................................................ | 29 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 |
| ..................GTTCAAGGATTGTCTGCAAAAC........................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................TCAAGGATTGTCTGCAAAACACCAGCA................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................TTGTCTGCAAAACACCAGCAT................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................TCAAGGATTGTCTGCAAAACACCAG................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TTTTATGACGTCAAGGCGTTGG.................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........GTCAGACGTTCAAGGATTGTCG............................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................CATTACTCCGAAGGTAAA............................................. | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............CTGTCGTTCAAGGATGGTCT............................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................TTCTCAGGGTTTGTA.................................................................................. | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................TTCATTTTTCTGAAGT.................................................................... | 16 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGGCAGGTGAGTGGTTTTA..................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GTACCTCCCCACAGTCTGCAAGTTCCTAACAGACGTTTTGTGGTCGTAACCATTTATCAAAGAAAGAGACCCAAACATGTAAAAAGACTTCAAAAATGTAAAGAGGCGTCCATTTACCAAAATACTGCAGTTCCGCAACCACCTCTTCTTAGCACCCGTC
**************************************************.................(((((..((((.(((((...)))))...)))).....)).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................CTTCAAAAATGTGAAGAGGCGT................................................... | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ........CCACAGTCTGCAAGTTCCTA.................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ....................................................TTCATCAAAGAAAGAG............................................................................................ | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| .....TCCCCACAGTCGCCAAGTC........................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 02:53 AM