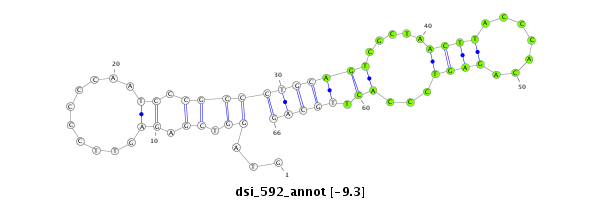
ID:dsi_592 |
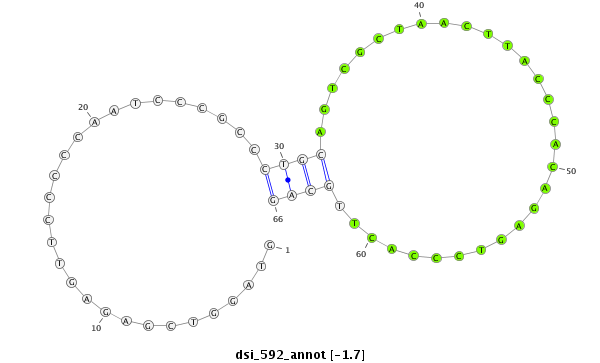
Coordinate:3l:8151103-8151168 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.7 | -9.3 | -9.3 | -9.3 |
 |
 |
 |
 |
CDS [3l_8150216_8151102_-]; exon [3l_8149985_8151102_-]; CDS [3l_8151169_8151873_-]; exon [3l_8151169_8151873_-]; intron [3l_8151103_8151168_-]
No Repeatable elements found
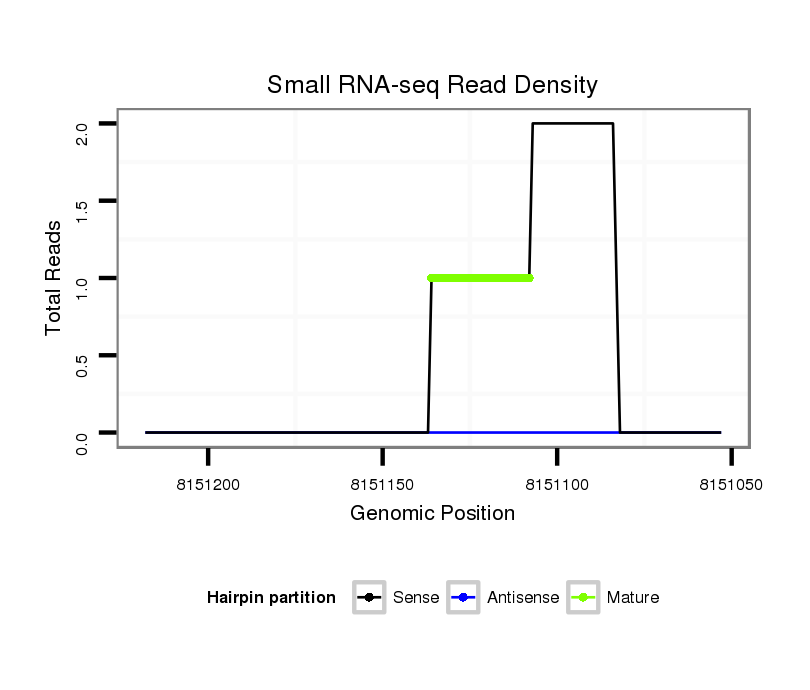
| ##################################################------------------------------------------------------------------################################################## ACACGTACAGACGATGGCCCAGCGAATGCTCCTCGCCACGGCTATTTGTGGTAGGTCGAGAGTTCCCCCAATCCCGCCCTGCAGTCGCTAACTTACCCACAGAGTCCCACTTGCAGGCACTGCAGGCACGTCTCTCCCGGCAGCAAAGGAGTCAGGGCAGCAGCGG **************************************************............................((((..............................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
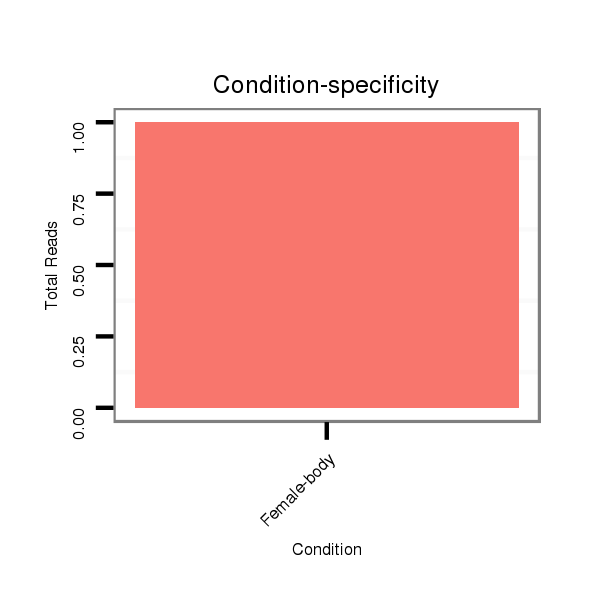
M053 female body |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
M023 head |
GSM343915 embryo |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................CAGCAAATGATTCAGGGC........ | 18 | 2 | 5 | 2.00 | 10 | 6 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 |
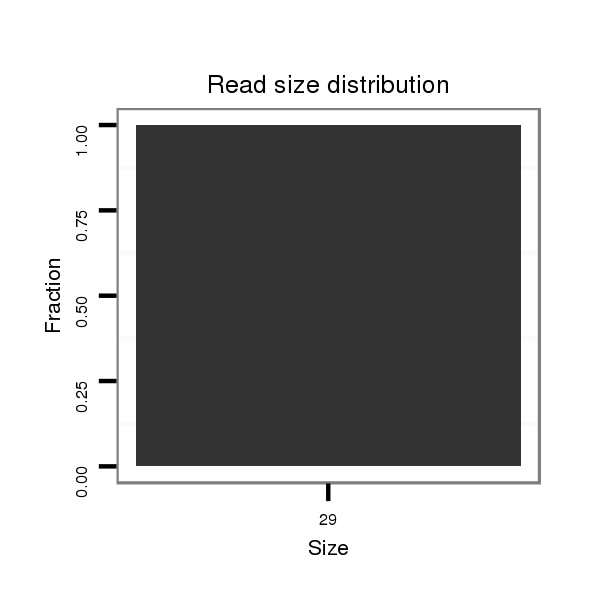
| ..................................................................................AGTCGCTAACTTACCCACAGAGTCCCACT....................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TGCAGGCACTGCAGGCACGTCTCT............................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TGCAGGCACTGCAGGCACGTCTCTC.............................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................GTCTATTTGTGGAAGATCGAGA......................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AATTCCACCAGTCCCGCCCTGC.................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................GTAGGTCGCGAGTTC..................................................................................................... | 15 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................TTTGTTGTGGGTCGAGAGTT...................................................................................................... | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................AGCCCCGTCTCTCGCGGCA........................ | 19 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AGGTCGAGAGTTCGCCT................................................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................TGTCGTAGGTCGAGA......................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................AGATCGAGAGTTCCCC.................................................................................................. | 16 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................AGCGAAGGCTCCTCGT.................................................................................................................................. | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GGAGGTCGAGAGTTC..................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................AGCGAAGGCTCCTCGTC................................................................................................................................. | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................TATAGGTCGAGAGTTC..................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................GGTCGAGAGTTCGCGC................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TGTGCATGTCTGCTACCGGGTCGCTTACGAGGAGCGGTGCCGATAAACACCATCCAGCTCTCAAGGGGGTTAGGGCGGGACGTCAGCGATTGAATGGGTGTCTCAGGGTGAACGTCCGTGACGTCCGTGCAGAGAGGGCCGTCGTTTCCTCAGTCCCGTCGTCGCC
**************************************************............................((((..............................))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|
| ..................................................................................................TGGCTCAGGGTCAACGTCC................................................. | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...............................................................AGAGGGTTAGTTCGGGACG.................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ...........................................................................................................GTCAACGGCCGTGACGTC......................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 |
Generated: 04/24/2015 at 10:43 AM