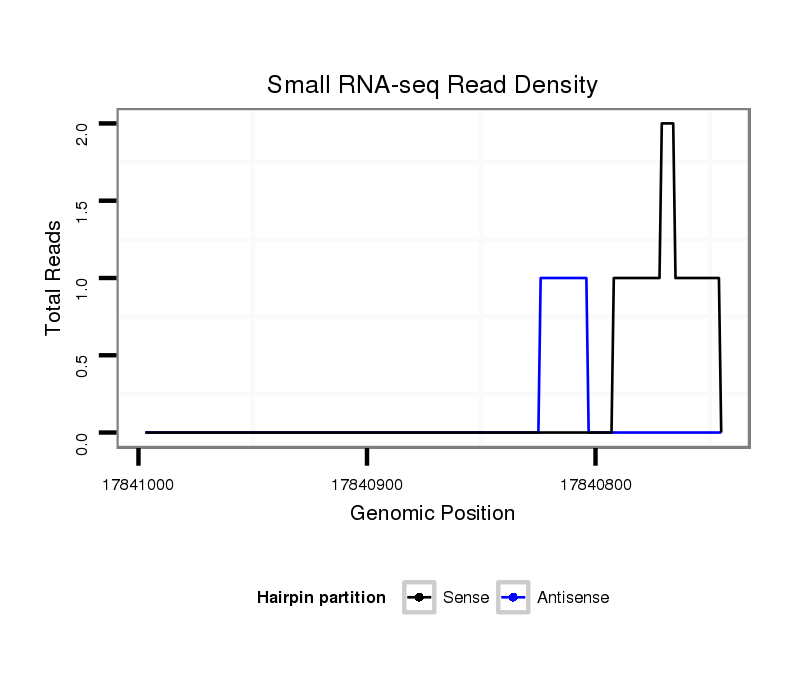
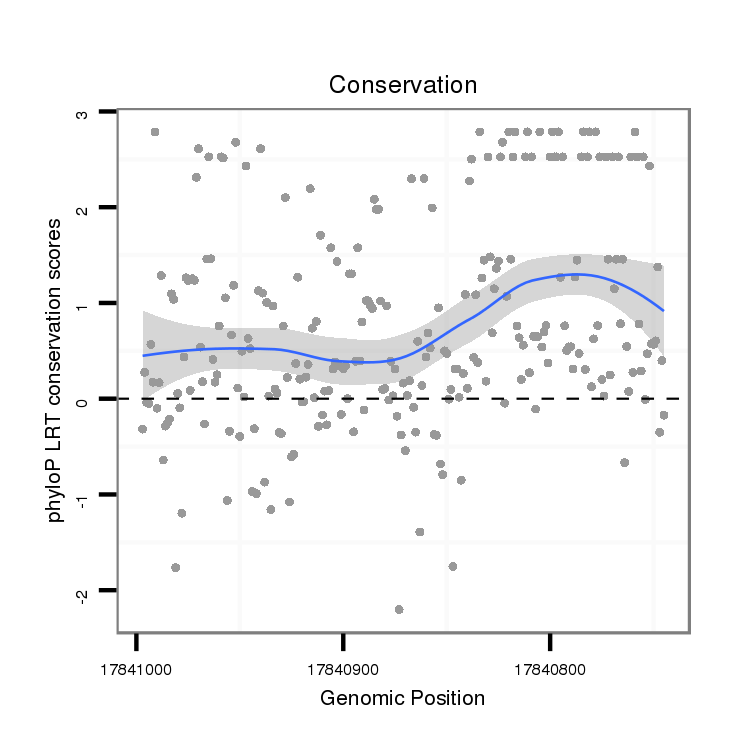
| AATTGCAGCAA-----------------------------------CAACA---------ACAAC--------------------AGCAGCAACA---ACAACAGC--------------------AGCAGCAACAA------CAGCATCAGC-----------------------------------AACAGCAAC---AGCAGCAACGAAT---------------------------------------------------------GCAACAGCAACAGCAACAGCAGCAACAAT------------T------GCA---GC----------------------------AAC--AACAACAACAGCAACA------------ACAACAAC--------------------------------------------AGCAGCAACAACAACAG---CAGCAGCAACA------ACAGCAACAACAACA---------------GCAGCAGCAACAGCAACAACAGCAGCA--GCA------------------------ | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| AATTGCAGCAA-----------------------------------CAACA---------ACAAC--------------------AGC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................AAT------------T------GCA---GC----------------------------AAC--AACAACAACAGC............................................................................................................................................................................................... | 24 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................T------GCA---GC----------------------------AAC--AACAACAACAGCAACA------------ACA............................................................................................................................................................................ | 28 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAACAACAACA---------------GCAGCAGC................................................ | 20 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ......................................................................................GCAGCAACA---ACAACAGC--------------------A................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| ......................................................................................................................................................................................................................................................................................AACAGCAACAGCAGCAACA......................................................................................................................................................................................................................................................................... | 19 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 |
| .............................................................................................................................................................................................................CAACGAAT---------------------------------------------------------GCAACAGCAACAGCAAC................................................................................................................................................................................................................................................................................... | 25 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................T------GCA---GC----------------------------AAC--AACAACAACAGCAACA------------............................................................................................................................................................................... | 25 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................TCAGC-----------------------------------AACAGCAAC---AGCAGCAACGAA.............................................................................................................................................................................................................................................................................................................................................................. | 26 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................AAT------------T------GCA---GC----------------------------AAC--AACAACAACAG................................................................................................................................................................................................ | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................ACAACAGC--------------------AGCAGCAACAA------C.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| .....................................................................................................................................................................................................................................................................................................AACAAT------------T------GCA---GC----------------------------AAC--AACAACAACAGC............................................................................................................................................................................................... | 27 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAG---CAGCAGCAACA------AC.................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| AATTGCAGCAA-----------------------------------CAACA---------ACAAC--------------------AG........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................AGC-----------------------------------AACAGCAAC---AGCAGCAACG................................................................................................................................................................................................................................................................................................................................................................ | 22 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................CAACAA------CAGCATCAGC-----------------------------------AACAGCAAC---AG........................................................................................................................................................................................................................................................................................................................................................................ | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................CAA------CAGCATCAGC-----------------------------------AACAGCAAC---AGCAG..................................................................................................................................................................................................................................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................CAACAGC--------------------AGCAGCAACAA------CAGCATCAG.......................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| AATTGCAGCAA-----------------------------------CAACA---------ACA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................AAT------------T------GCA---GC----------------------------AAC--AACAACA.................................................................................................................................................................................................... | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................CAACAA------CAGCATCAGC-----------------------------------AACAGC................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAG---CAGCAGCAACA------ACAGCAACAA............................................................................ | 25 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................AGCAACAGCAACAGCAGCAACAAT------------T------.................................................................................................................................................................................................................................................... | 25 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................T---------------------------------------------------------GCAACAGCAACAGCAACAGCAGC............................................................................................................................................................................................................................................................................. | 24 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................AGCATCAGC-----------------------------------AACAGCAAC---AGCAG..................................................................................................................................................................................................................................................................................................................................................................... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................CAA------CAGCATCAGC-----------------------------------AACAGC................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................ATCAGC-----------------------------------AACAGCAAC---AGCAGCAAC................................................................................................................................................................................................................................................................................................................................................................. | 24 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAACA------ACAGCAACAACAACA---------------GCA..................................................... | 26 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAGCAACAACAGCAGCA--G.......................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGCAGCAACAGCAACAACAGCAG............................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GCAGCAACAGCAACAACAGCAGCA--GC......................... | 26 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACA---------------GCAGCAGCAACAGCA......................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACA------ACAGCAACAACAACA---------------GCAGCA.................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................CAACAGCAACAGCAACAGCAG.............................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................CAACA---ACAACAGC--------------------AGCAGC.............................................................................................................................................................................................................................................................................................................................................................................................................................................. | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................AACAAT------------T------GCA---GC----------------------------AAC--AACAACAG................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................A---ACAACAGC--------------------AGCAGCAACAA------................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAG---CAGCAGCAACA------ACAGCAACA............................................................................. | 25 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAACA---------------GCAGCAGCAACA............................................ | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAACA---------------GCAGCAGCAACAG........................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................AC--------------------AGCAGCAACA---ACAACAGC--------------------A................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAACA---------------GCAGCAGCAA.............................................. | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CA---------------GCAGCAGCAACAGCAACAA..................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAACAG---CAGCAGCAACA------A..................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................AGC-----------------------------------AACAGCAAC---AGCAGCA................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................ACAAC--------------------------------------------AGCAGCAACAACAACA........................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................A------ACAGCAACAACAACA---------------GCAGCAGCA............................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................................................................................................AGCAACAGCAGCAACAAT------------T------G................................................................................................................................................................................................................................................... | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................AACAACAACAGCAACA------------ACAACA......................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................ACA------------ACAACAAC--------------------------------------------AGCAGCAA................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAACAACA---------------GCAGCAGCAA.............................................. | 19 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACA------ACAGCAACAACAACA---------------GCAGCA.................................................. | 26 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAACAG---CAGCAGC................................................................................................ | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAG---CAGCAGCAACA------ACA................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................................................................AACAGCAACAGCAACAGCAGC............................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |