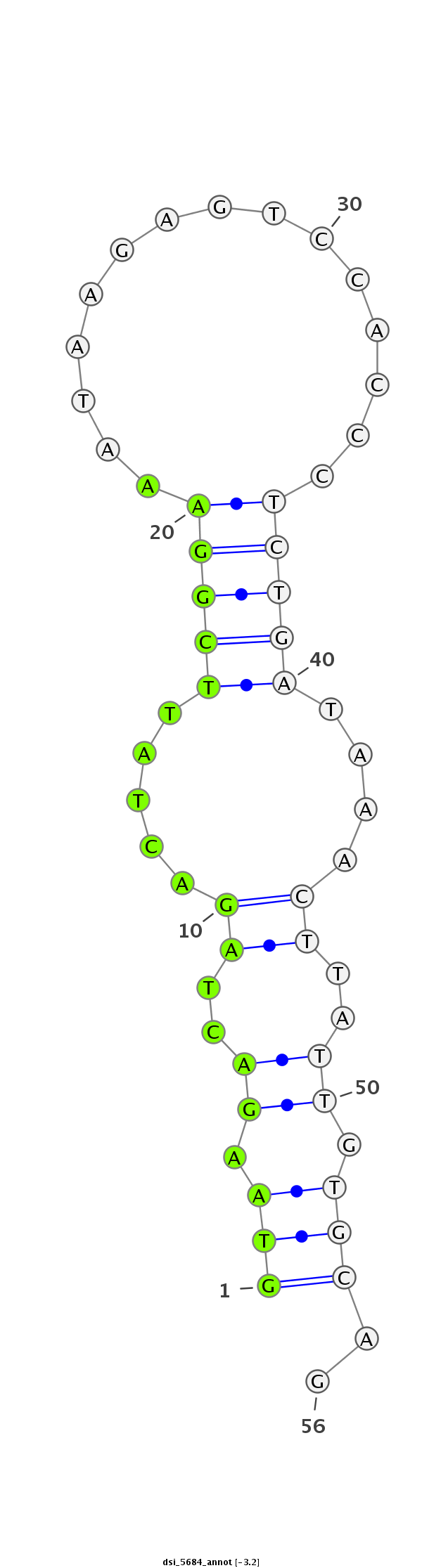
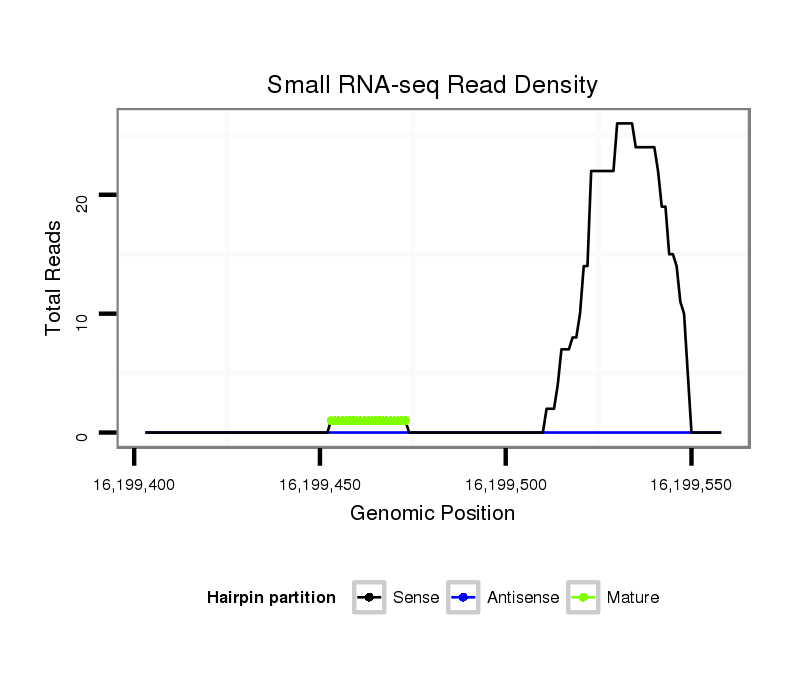
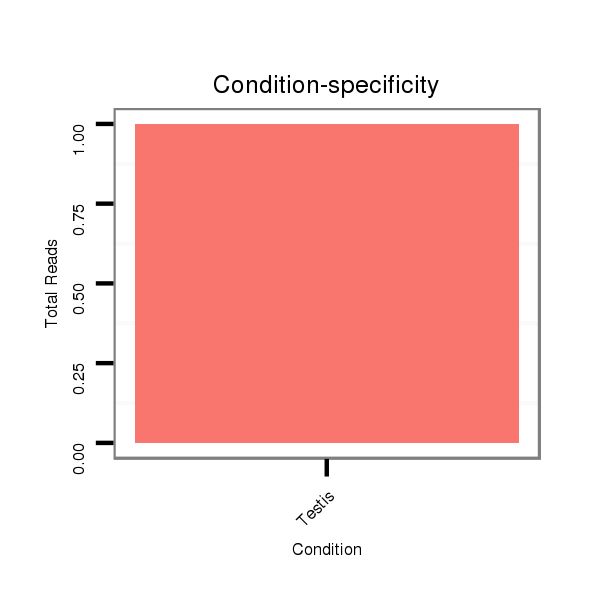
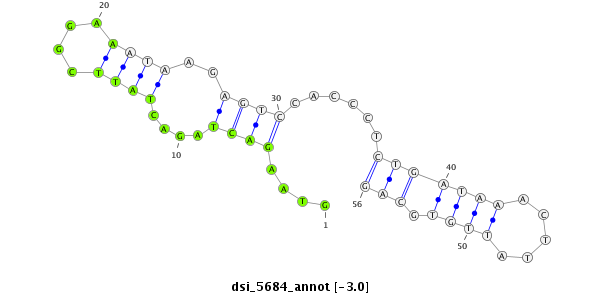
ID:dsi_5684 |
Coordinate:3l:16199453-16199508 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -3.0 | -3.0 | -2.9 |
 |
 |
 |
CDS [3l_16199509_16200631_+]; CDS [3l_16199343_16199452_+]; exon [3l_16199509_16200631_+]; exon [3l_16199509_16200631_+]; CDS [3l_16199343_16199452_+]; CDS [3l_16199509_16200631_+]; exon [3l_16199343_16199452_+]; exon [3l_16199343_16199452_+]; intron [3l_16199453_16199508_+]; intron [3l_16199453_16199508_+]
No Repeatable elements found
| ##################################################--------------------------------------------------------################################################## GAGAATCTGGATGAGGAAATTGTTCCTTATTTACCCACACTTATGGATCGGTAAGACTAGACTATTCGGAAATAAGAGTCCACCCTCTGATAAACTTATTGTGCAGTCTGTTTGGGGTTATGGAACCGCAGAACACGAATCGAATGCGTGAAATGG **************************************************(((.((..((.....(((((...............)))))....))..)).)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GCAGAACACGAATCGAATGC......... | 20 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGGAACCGCAGAACACGAATCGAATG.......... | 26 | 0 | 1 | 3.00 | 3 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TGGGGTTATGGAACCGCAGAACACGAA................. | 27 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| ............................................................................................................TGTTTGGGGTTATGGAACCGCAGA........................ | 24 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ......................................................................................................................TATGGAACCGCAGAACACGAATCGAATG.......... | 28 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGGAACCGCAGAACACGAATCGAA............ | 24 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TTATGGAACCGCAGAACACGAATC............... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................ATTTACCCACACTTATGGATCCTC........................................................................................................ | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................TTGGGGTTATGGAACCGCAGAACACGA.................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGACTAGACTATTCGGAA..................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................TGGAACCGCAGAACACGAATCGAATGC......... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................TTGGGGTTATGGAACCGCAGAACACGAA................. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TGGGGTTATGGAACCGCAGAACACGA.................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GGTTATGGAACCGCAGAACACGAATC............... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TATGGAACCGCAGAACACGAATCGAA............ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGGAACCGCAGAACACGAATCGA............. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGGAACCGCAGAACACGAATCGAAT........... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................TATGGAACCGCAGAACACGAATC............... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................ACGAATCTAACGCGTGA..... | 17 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................TTATGGAACCCCAGTAC...................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................ATCGAATGCGTGAAG... | 15 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CGAAGCTAATGCGTGA..... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................TTATGTATCGGGAAGATTA................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CTCTTAGACCTACTCCTTTAACAAGGAATAAATGGGTGTGAATACCTAGCCATTCTGATCTGATAAGCCTTTATTCTCAGGTGGGAGACTATTTGAATAACACGTCAGACAAACCCCAATACCTTGGCGTCTTGTGCTTAGCTTACGCACTTTACC
**************************************************(((.((..((.....(((((...............)))))....))..)).)))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|
| .............................................................................CAGGTGGGAAACTAT................................................................ | 15 | 1 | 7 | 0.14 | 1 | 1 | 0 |
| ....................................................................................................................................TGTGCTAAGCTTGCACACT..... | 19 | 3 | 16 | 0.13 | 2 | 0 | 2 |
Generated: 04/24/2015 at 02:52 AM