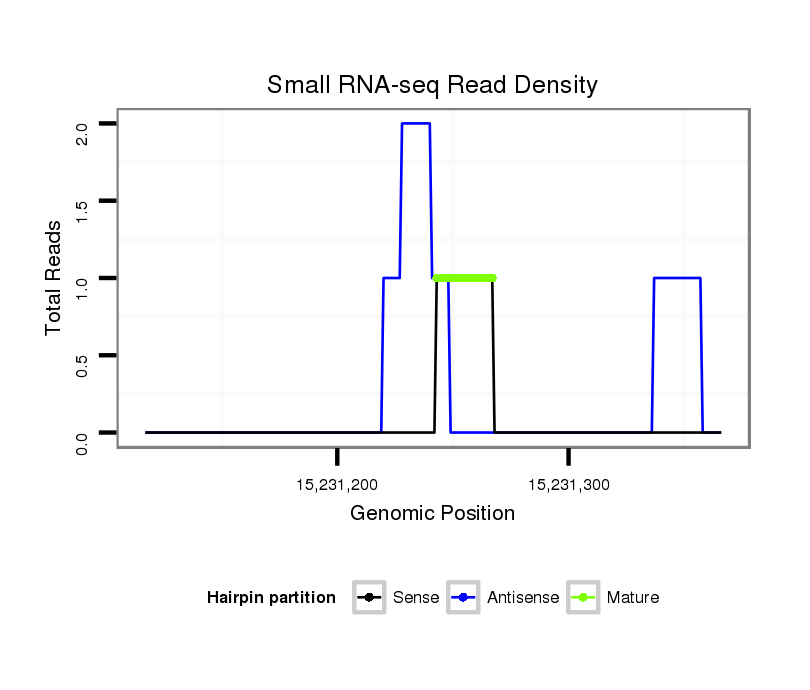
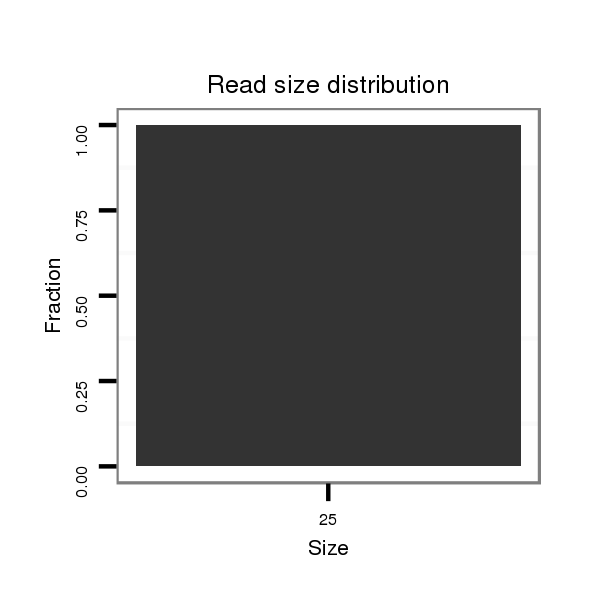
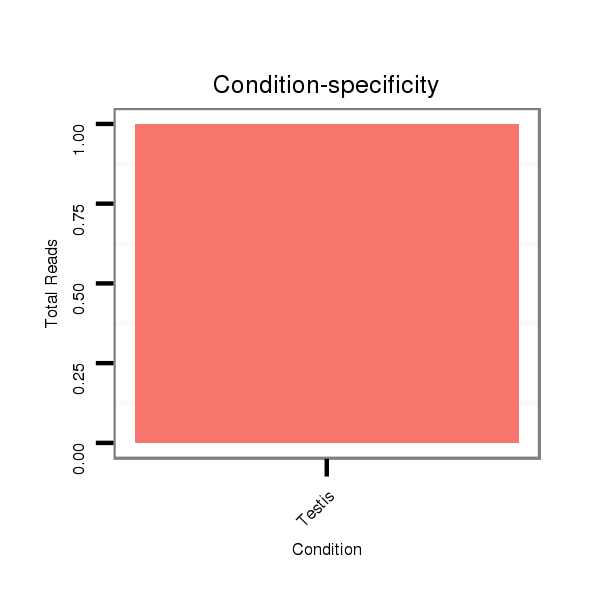
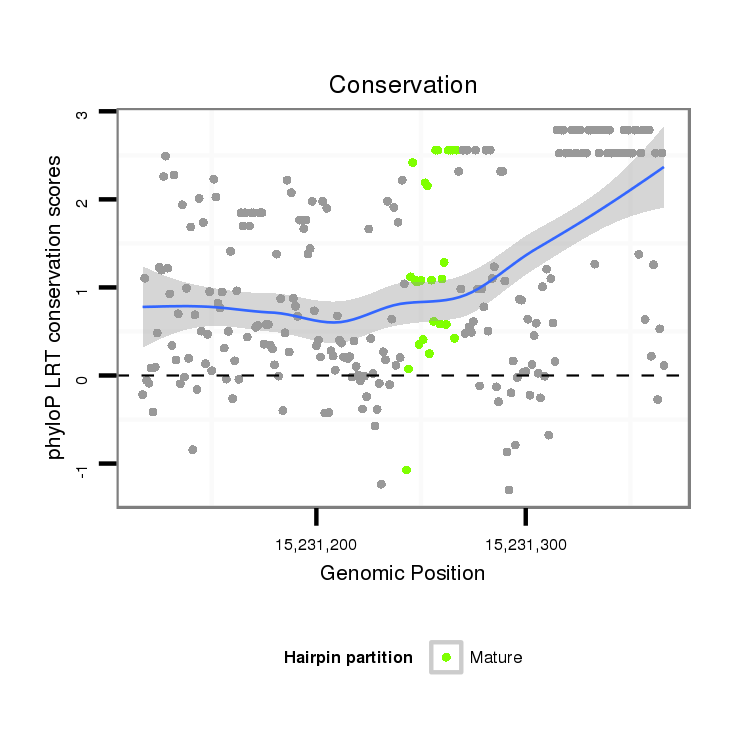
| ..............................................................GAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
19 |
2 |
2 |
2232.50 |
4465 |
3264 |
816 |
274 |
59 |
3 |
6 |
21 |
4 |
11 |
4 |
3 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
7 |
2121.29 |
14849 |
7968 |
1488 |
3564 |
704 |
567 |
341 |
104 |
94 |
1 |
1 |
0 |
13 |
0 |
3 |
1 |
| ...............................................................AGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
18 |
2 |
5 |
863.60 |
4318 |
3430 |
784 |
47 |
27 |
5 |
5 |
6 |
2 |
5 |
2 |
2 |
0 |
3 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTTGTGGA........................................................................................................................................................................ |
21 |
3 |
2 |
46.50 |
93 |
0 |
0 |
24 |
58 |
9 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
20 |
2 |
1 |
34.00 |
34 |
0 |
0 |
0 |
26 |
5 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTTGTG.......................................................................................................................................................................... |
19 |
2 |
3 |
10.33 |
31 |
0 |
0 |
0 |
24 |
4 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGAATGTTGTGGT........................................................................................................................................................................ |
21 |
3 |
2 |
9.50 |
19 |
0 |
0 |
18 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGAATGTTGTGGT........................................................................................................................................................................ |
20 |
2 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGACTGTTGTGG......................................................................................................................................................................... |
20 |
3 |
10 |
1.50 |
15 |
0 |
0 |
15 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGTATGTTGTGG......................................................................................................................................................................... |
20 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGATTGTTGTGGA........................................................................................................................................................................ |
21 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................AGTTGGTGTATGTTGTGG......................................................................................................................................................................... |
18 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................AGGAGTTGGTGAATGTTGTGGT........................................................................................................................................................................ |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................................................................................TTGTCGCGGCTCAGGTAAGTC......... |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGTATGTTGTGGA........................................................................................................................................................................ |
21 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................GGGTCTGGAGGCTTACTTTGT.............................................................................................................................. |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................CGCCGAGTTGGTGAATGTTGTG.......................................................................................................................................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................GCCGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................CGGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................CCGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
21 |
3 |
2 |
1.00 |
2 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................CCGAGTTGGTGAATGTTGTG.......................................................................................................................................................................... |
20 |
3 |
6 |
1.00 |
6 |
0 |
0 |
1 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGATTGTTGTG.......................................................................................................................................................................... |
18 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGAATGTTGTGGTA....................................................................................................................................................................... |
21 |
3 |
2 |
1.00 |
2 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................GGTTGTAAATGAACGAGATGGT..................................................................................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................AGGCTTACTTTGTTTACTTGA...................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGCATGTTGTG.......................................................................................................................................................................... |
19 |
3 |
14 |
0.79 |
11 |
0 |
0 |
11 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................AGAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
10 |
0.70 |
7 |
0 |
0 |
2 |
4 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTTGTGGAT....................................................................................................................................................................... |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGCATGTTGTGGA........................................................................................................................................................................ |
20 |
3 |
4 |
0.50 |
2 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CGCAGGTGTTTGTTGTGGTTGT..................................................................................................................................................................... |
22 |
3 |
4 |
0.50 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................AAAAGAACCGGGTTGGGATA.................................................................... |
20 |
2 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................AAAAAACAGGCTTGGGAT..................................................................... |
18 |
2 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGCATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
3 |
0.33 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................TTGGGATAGAGGCTT............................................................. |
15 |
1 |
4 |
0.25 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..........................................................CGCCGAGTTGGTGAATGTTG............................................................................................................................................................................ |
20 |
3 |
4 |
0.25 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................TTGGTATACAGGCCTGG........................................................... |
17 |
2 |
10 |
0.20 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGGATGTTGTGG......................................................................................................................................................................... |
19 |
2 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTTGT........................................................................................................................................................................... |
18 |
2 |
11 |
0.18 |
2 |
0 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGGGTTGGTGATTGTTGTGGA........................................................................................................................................................................ |
21 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................................TTGGGATAGAGGCTTGT........................................................... |
17 |
2 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................TTGTGGTAGTAAATGTATGA........................................................................................................................................................... |
20 |
3 |
17 |
0.12 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
| ..........................................................................................................................................................................TGGGAGGGGATACAGGCT.............................................................. |
18 |
2 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................AGGAGTTGGTGAATGTTGTG.......................................................................................................................................................................... |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................TTGATGAGGGTCTGTAGGTT...................................................................................................................................... |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ...................................................................................................................................................................................................AAATGAGCCGTGGTAGGCA.................................... |
19 |
3 |
10 |
0.10 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGAATGTTGTGGG........................................................................................................................................................................ |
20 |
3 |
10 |
0.10 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CGAGTTGGTGGATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
20 |
0.10 |
2 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTGGTGAATGTAGTGG......................................................................................................................................................................... |
20 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...CGTCGTTGTTGTTGTAGGTTGTTC............................................................................................................................................................................................................................... |
24 |
3 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........TTGGTGTCGGTTGGTT............................................................................................................................................................................................................................... |
16 |
1 |
12 |
0.08 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GTAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
12 |
0.08 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGTGAGTGTTGTGGA........................................................................................................................................................................ |
20 |
3 |
13 |
0.08 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................GGAGTTTGTGAATGTTGTGG......................................................................................................................................................................... |
20 |
3 |
14 |
0.07 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................AAAAAACTAGGTTGG........................................................................ |
15 |
1 |
15 |
0.07 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| .............................................................CGAGTTGGTGAGTGTTGTGG......................................................................................................................................................................... |
20 |
3 |
16 |
0.06 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................CGGCTCAAGTAAGTC......... |
15 |
1 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..............................................................................................................................................................................TTGGGATCGAGGCTTG............................................................ |
16 |
2 |
19 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ..............................................................GAGTTGGGGAATGTTGTGG......................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................................................................................AAAAAATGAAAAAAGCTGGGT........................................................................... |
21 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................TAGTTGGTGAATGTTGTGG......................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................AGCGGCTCAGGTAAGAC......... |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................AAAAAAACTGGGTCG......................................................................... |
15 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................GAGCTGGTGAATGTTGTGG......................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |