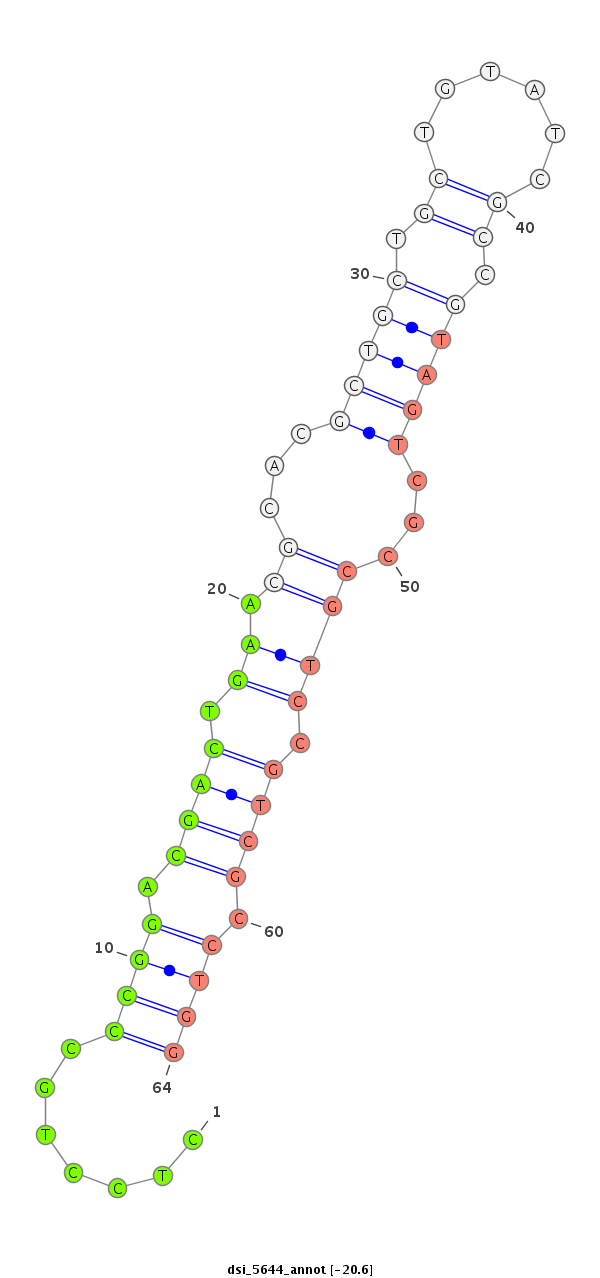
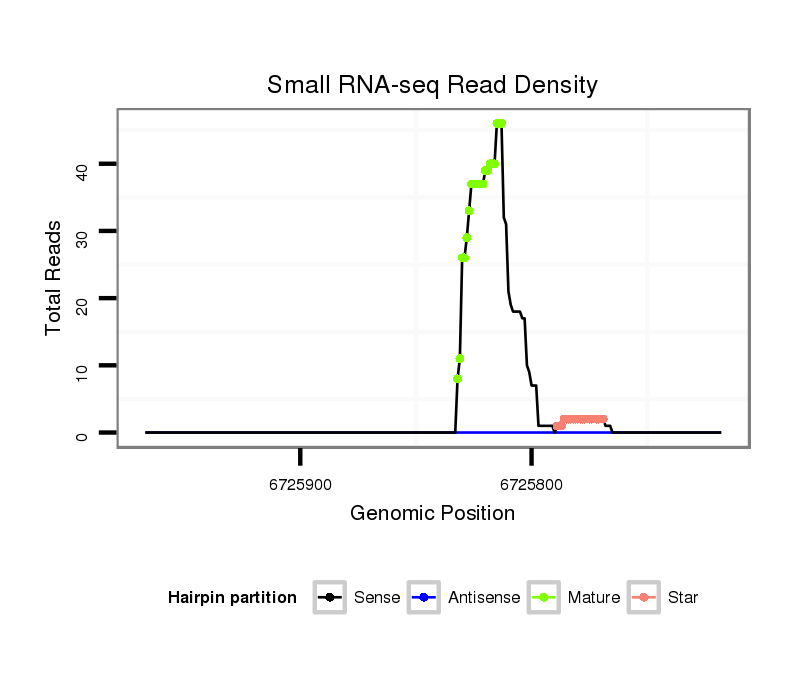

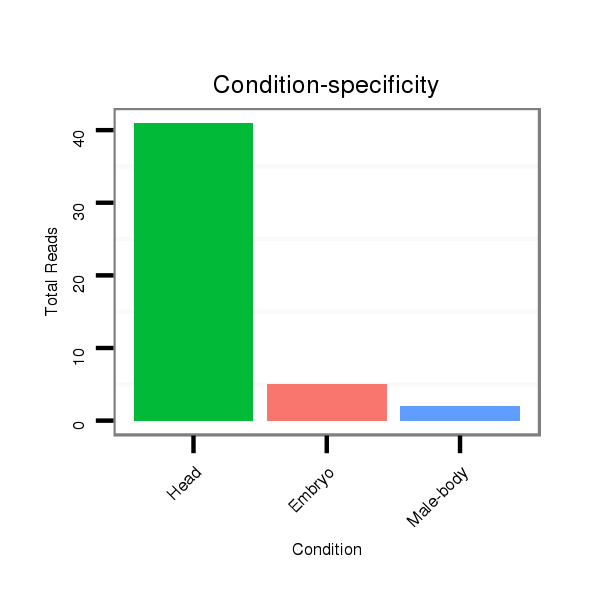
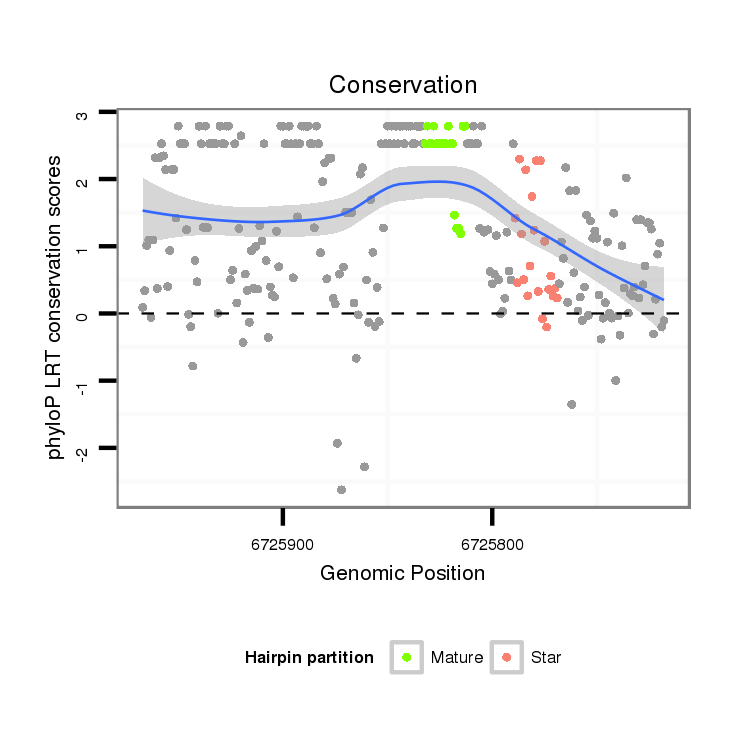
ID:dsi_5644 |
Coordinate:3r:6725768-6725917 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.6 | -20.5 | -19.7 |
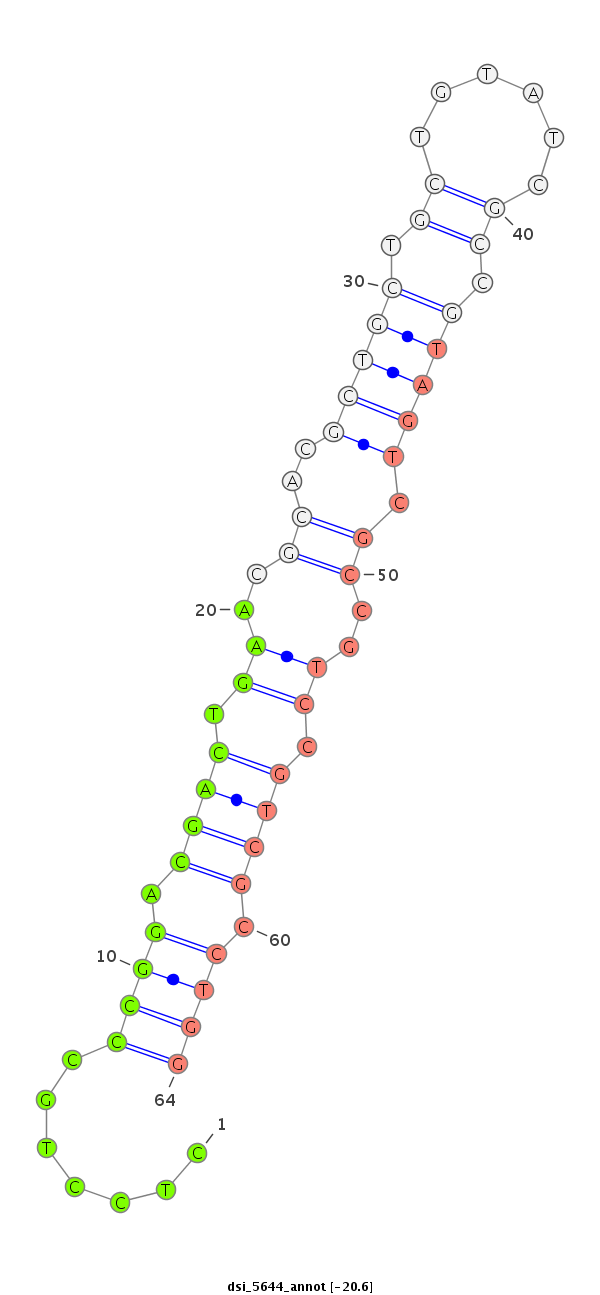 |
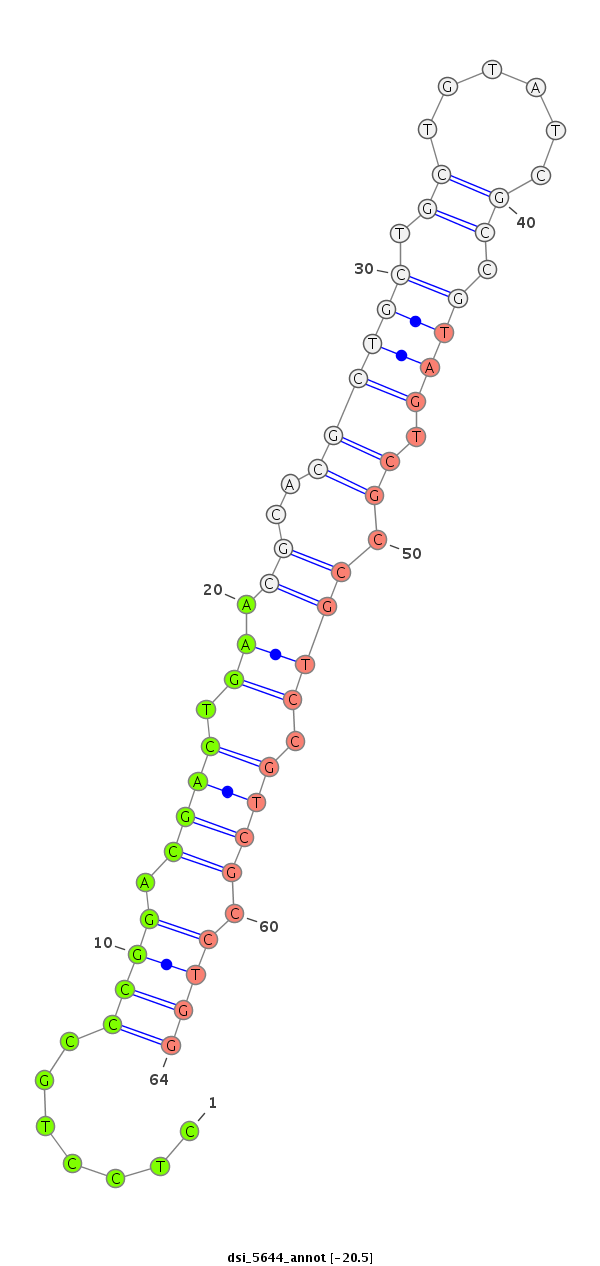 |
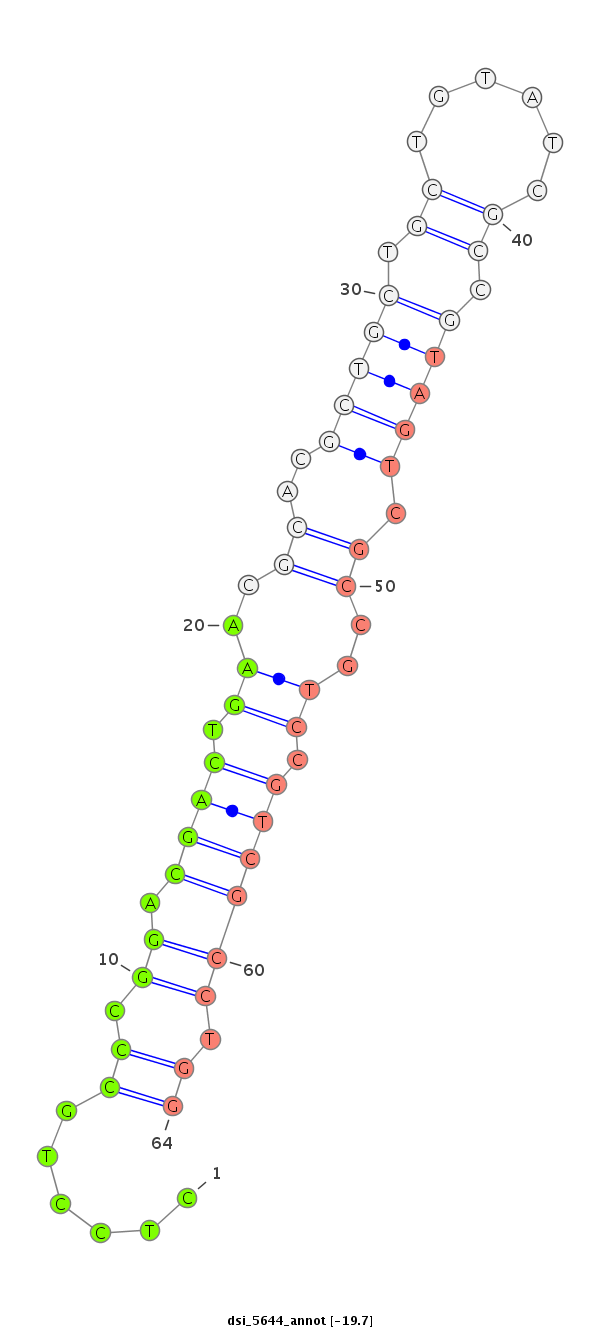 |
five_prime_UTR [3r_6725429_6725833_-]; exon [3r_6725251_6725833_-]; exon [3r_6725251_6725904_-]; five_prime_UTR [3r_6725251_6725904_-]
No Repeatable elements found
| ---------------------------------------------------------------########################################################################################################################################################################################### GCGGAGGCGCAGGCGCATGCGCAGATGTCCTGCGCCATGGAAGGCGCAAGGTGGGGAGCTCCAGCCTTCCAGGCTGAATACCTACTGCTGCTCGACGCTTTTTGGCGAGCCGTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTGCTGCTGTATCGCCGTAGTCGCCGTCCGTCGCCTGGAAAAAAGAACTCCCCGGAGCTACATTGTATAGCCAGGAATAGAAAAAAAAA ***************************************************************************************************************************************.......((((.((((.((.((...(((((.((......)).)))))...)))).)))).))))*************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553487 NRT_0-2 hours eggs |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
O002 Head |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................ATGCCTACTGCTGCTAGACT......................................................................................................................................................... | 20 | 3 | 2 | 15.50 | 31 | 0 | 30 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................ATGCCTACTGCTGCTAGACTC........................................................................................................................................................ | 21 | 3 | 1 | 13.00 | 13 | 0 | 9 | 0 | 3 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................CTCCTGCCCGGACGACTGAA............................................................................................... | 20 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................GAACGCACGCTGCTGCTG................................................................................ | 18 | 0 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCTGCCCGGACGACTGAACG............................................................................................. | 20 | 0 | 1 | 6.00 | 6 | 5 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCTGCCCGGACGACTGAA............................................................................................... | 18 | 0 | 1 | 6.00 | 6 | 5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................CCCGGACGACTGAACGCACGCTGC..................................................................................... | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TGCCCGGACGACTGAACG............................................................................................. | 18 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CGACTGAACGCACGCTGC..................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTCCTGCCCTGACGACTGAAC.............................................................................................. | 21 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TCCTGCCCGGACGACTGAA............................................................................................... | 19 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................CTCCTGCCCGGACGACTGAACG............................................................................................. | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GCCCGGACGACTGAACGCACGCTGC..................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GCCCGGACGACTGAACGC............................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................CCCGGACGACTGAACGCACGCTGCTG................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCTGCCCGGACGACTGAACGCACGCTGC..................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................TAGTCGCCGTCCGTCGCCTGG................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TAGTCGCCGTCCGTCGCCTGGG.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GCCCGGACGACTGAACGCACGCTGCT.................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GCCCGGACGACTGAACGCA........................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TCCTGCCCGGACGACTGAAC.............................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCTGCCCGGACGACTGAACGCACGCT....................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................CACGCTGCTGCTGTATCGCC......................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ACTGAACGCACGCTGCTG................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................CCTGCCCGGACGACTGAACGC............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................TCGCCGTCCGTCGCCTGGAAA................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................CGAGCCGTCGTGAGACAG............................................................................................................................... | 18 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................GTCGAGCCGTCGTGAGACAG............................................................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................CCTTCCAAGCTGAATA.......................................................................................................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................TGCACAGATGTCCTGGGC....................................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................CGAGCTGTCGTGAGACAG............................................................................................................................... | 18 | 3 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| ..............................TGCGCCAGGGAAGGCCC........................................................................................................................................................................................................... | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................GCAAGCCGTCGTGAGACAG............................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................CTTCCAAGCTGAATA.......................................................................................................................................................................... | 15 | 1 | 17 | 0.12 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................AGCCTTCCAAGCTGAATGTC........................................................................................................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................CCTTCCAAGCTGAATACGA....................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................................TGCCTACTGCTGCTAGACT......................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................TGGGCAGCTCCATCCTTC..................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................CACGTAAGGCGCAAGG....................................................................................................................................................................................................... | 16 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................GAGCCGTCGTGAGACAGG.............................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................CCTGTAAGGCGCAAGG....................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................CCGGGTGGGGAGCTCCA........................................................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CCGTCGTGAGACAGAACT........................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CGCCTCCGCGTCCGCGTACGCGTCTACAGGACGCGGTACCTTCCGCGTTCCACCCCTCGAGGTCGGAAGGTCCGACTTATGGATGACGACGAGCTGCGAAAAACCGCTCGGCAGCGGTCTGTCTTCAATGGAAACGAGGACGGGCCTGCTGACTTGCGTGCGACGACGACATAGCGGCATCAGCGGCAGGCAGCGGACCTTTTTTCTTGAGGGGCCTCGATGTAACATATCGGTCCTTATCTTTTTTTTT
***************************************************.......((((.((((.((.((...(((((.((......)).)))))...)))).)))).))))*************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................ACGGATGACGACGATCTGAG........................................................................................................................................................ | 20 | 3 | 4 | 1.25 | 5 | 4 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................CGGCAGGCAGCGTACCA.................................................. | 17 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................GGTCGGACGGCCCGAGTTA........................................................................................................................................................................... | 19 | 3 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................TCTTTAATGGAGCCGAGGA.............................................................................................................. | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................CGTACGGATGACGACGATC............................................................................................................................................................ | 19 | 3 | 18 | 0.11 | 2 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................GCCCTCGTTGTATCATAT.................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:6725718-6725967 - | dsi_5644 | GCGGAGGCGCAGGCGCATGCGCAG--ATGTCCTGCGCCATGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T---------CCAG-CCTTCCAGGCT---------------GAATACCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--CTGCTC-------------G-AC-----GCTTTTTGGC-----GA--GCC-----GTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTA---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAA-A---AA-------------------------------------------------AGAACTCCC----------CGGAGCTACATTGTA-TAGCCAGG--------------AATAGAAAAAAAAA-------- |
| droSec2 | scaffold_5:5369470-5369717 - | GCGGAGGCGCAGGCGCATGCGCAG--ATGTCCTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T---------CCAG-CCTTCCAGGCT---------------GAATACCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--CTGCTC-------------A-AC-----GCTTTTTGGC-----AA--GCC-----GTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTA---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAA-----AA-------------------------------------------------AGAACTCCC----------CGGAGCTACATTGTA-TAGCCAGG--------------AATAGAAAA-AAAA-------- | |
| dm3 | chr3R:14531111-14531367 + | GCGGAGGCGCAGGCGCATGCGCAG--ATGTCCTGCGCCATGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T---------CCAG-CCTTCCAGGCT---------------GAATACCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--CTGCTC-------------G-AC-----GCTTTTTGGC-----CA--GCC-----GTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTA---ACGCCGTAGTC--------GCCGTCCGTCGCCTGGAA-----AA-------------------------------------------------AGAACTCCC----------CGGAGCTACATTGTA-TAGCCAGG--------------AATAGAAAAAAAAAAAAAATAA | |
| droEre2 | scaffold_4770:7102111-7102350 - | GCGGAGGCGCAGGCGCATGCGCAG--ATGTCCTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T---------CCAG-CCTTCCAGGCT---------------GAATACCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--CTGCTC-------------T-GC-----GCTTTTCGGC-----GA--GCC-----GTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTA---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAA-----A--------------------------------------------------AGAACTCCC----------CGGAGCTACAT-GCA-TAGCCCGA--------------AATAGAA--------------- | |
| droYak3 | 3R:3189057-3189297 + | GCGGAGGCGCAGGGGCATGCGCAG--ATGTCTTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T---------CCAG-CCTTCCAGGCT---------------GAATACCTAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------TG--CTGCTC-------------T-GC-----GCTTTTCGGC-----CA--GCC-----GTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTA---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAA-----AA-------------------------------------------------AGAACTCCC----------CGGAGCTACAT-GTA-CAGCCAGA--------------AATAGAA--------------- | |
| droEug1 | scf7180000409804:760145-760379 + | GCGGAGGCGCAGGCGCATGCGCAT--ATGTCCTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------A----------CGG-CCTTCCAGGCT---------------GAATACCGACAAAA-A-----------------------------------------------------------------------------------TCGCTGAG--------------------------------------------------------------C-----TG--CTGCTC-------------T-GG-----GCTTTTTGGC-----GA--ACC----AGTTGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAACAAAA------T-----------------------------------------------------------------------------------------------------AATAGAAAAAAATA-------- | |
| droBia1 | scf7180000302402:257649-257932 + | GCGGAGGCGCAGACGCATGCGCAG--TTGTCCTACGCCCTGGAAAAGCGCA-----------------------CG-----------------------------------------------------GCGGGGAGC-------T----------CGG-CCTTCCAGGCT---------------GAATACCGACAAAAACGA--------------------------------------------------------------------------CG--------------------------------------------------------------------------------------AGCTCTGGCCTTTTCCACG-G-------------GGC-----TC--ACC-----ATCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGT------CTCGCCGCCCGTCGCCTGGAA-AAAA------T-ATAGAAA------------------AAATAATA----------AGAACTTCC----------CGGAGCAGCAGAGGAGCT-CCAGA-GCTC----TCGAAAATCG------AAA-------- | |
| droTak1 | scf7180000415206:17604-17900 - | GCGGAGGCGCAGGCGCATGCGCCT-CGAGTCCTGCGCCCTGGAA-AACGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T----------CGT-CATTCCAGGCT---------------GAATACCGACAAAAACGG--------------------------------------------------------------------------CG-------------------------------------------------------A---------GCT-CTGCT-CTGCTTTGCTCTGCTC-------------G-GG-----GCTTTTCAGCCAAGCGA--ACCAACCGTTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGT------CTCGCCGTCCGTCGCCTGGAA-----AAAGAAAAATAGAAATAGAAA------------A----GGA----------AAAACTCCC----------TGGAGC-----------T-CCAGA-GCTC----T-------AG-----AAAA-------- | |
| droEle1 | scf7180000490994:261768-262054 + | GCGGGGGCGCAGGCGCATGCGCAG--GTGTCCTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T----------CGG-CCTTCCAGGCT---------------GAATACCGACAAAA-CGA--------------------------------------------------------------------------CGA-ACGAC------------------------------------------CCG-----TGCAGT-AGCTACTGCTGC-----TG--CTGCTC-------------T-GC-----GCTTTTCAGCTCA--GA--ACC---CAGTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGCCTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGT------CTCGCCGTCCGTCGCCTGGAA-----A--------------------CCAAAAAAAATA--------AATAAATAAAATAAAAA--------------------------------------AATAAAAT------------AAAAAC-------- | |
| droRho1 | scf7180000779900:152768-153058 + | GCGGAGGCGCAGGCGCATGCGCAG--GTGTCCTGCGCCGTGGAA-GGCGCA-----------------------AG-----------------------------------------------------GTGGGGAGC-------T----------CGG-CCTTCCAGGCT---------------GAATACCGACAAAA-CGTC-------GAGCTGCTGCCGCTGCTGCTGCTGCTGCTGCTAGTGCTGGTG-----------------------------------------------------------------------------------------------------C-----TG--CTGCTC-------------T-GT-----GCTTTTCAGC-----GA--ACC---CAGTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGT------CTCGCCGTCCGTCGCCTGGAA-----A--------------------------------------------------AGAACTCCC----------CGGAGCAG------AGCT-CCAGA-GCC--------------G----AAATT-------- | |
| droFic1 | scf7180000453812:532185-532465 - | GCGGAGGCGCAGACGCATGCGCAG--ATGTCCTGCGCCGTGGAA-GGCGCA-----------------------CG-----------------------------------------------------TTGGGGACCA------T----------CGGCCCTTCCAGCCT-------------GTGAATACCGACAAAA-G-G--------------------------------------------------------------------------CG-------------------------------------------------------AGTGAGCTCAGCT-CTGCT-CTGCTTTGCTCTGCTC-------------T-GCAGTAGACTTTCTGGC-----GA--ACG-----CTCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCTGTT---TCGCCGTAGTC--------GCCGTCCGTCGCCTGGAAGAAAAAA-------------------------------------------------AAAACTCCC----------CGGAGCTGC--------T-CCAGA--------------ACT-G------AAA-------- | |
| droKik1 | scf7180000302256:488864-489139 - | GCGGTGGAGCAGGCGCATGCGCCA-TGTATCCTGCGCCATGGAA-GGCGCA-----------------------CGGC----------AGT-------------------------------------GGCGGGGAGCAGATCTCTCCGAACGGCACAA-CATTCCAGGCGCACTCCGTCGTCCATGAATACCGACAAAA-CG-------------------------------------------------------------------------AACGA-A----------------------------------------------GAGGCGAG-------AGCT-----------------CTGCCC------GTGGCGCT-GG-----GC---------------------------TGTCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACTGAACGCACGCTG-CTGCCGTTAG-TCGCCGTAGTC--------G------------TGGAA-----A----------------------------------------------------CACTCCC----------CGGAGCAG--------CTCCCAGA-GCC--------------C----AGTTC-------- | |
| droAna3 | scaffold_13340:6608166-6608449 - | ATGGTGGAGCAGGCGCCTGCGCAAACATATCCTGCGCCGTGGAA-AGCCCACACACACACAGATACAACGAGGAAG-----------------------------------------------------GTGGGGAGCA------T---------TCGG-CATTCCAGGCT---------------GAATACCGACAAAA-CAG--------------------------------------------------------------------------CATTTGCTTGCTGAGGGC-----------------------------------------------------------T-----TG--CTGCTC-------------G-CT-----GCTTTCTG----------------------GCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACAGAACGCACGCTG-TTGCCGCGT--CCGCTGGA---------------TCTGGGATCTGGGA-----A--------------------CTGGGAGAACTG------------------------------------------GTTTATTGCT-TTGCTGTGAACACCAA-C--------G---TAAAAC-------- | |
| droBip1 | scf7180000396708:2230764-2231038 + | ATGGTGGCGCAGGCGCCTGCGCAAACATATCCTGCGCCGTGGAA-AGCCCACACACAC----ATACGATGAGGAAG-----------------------------------------------------GTGGGGAGCA------T---------TCGG-CATTCCAGGCT---------------GAATACCGACAAAA-CAG--------------------------------------------------------------------------CAT----TTGCTGAGGGC-----------------------------------------------------------T-----TG--CTGCTC-------------G-CT-----GCTTTCTG----------------------GCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACAGAACGCACGCTG-CTGCCGCGT--CCGCTGGA---------------TCTGGGATCTGGGA-----A--------------------CTGGGAGAACTG------------------------------------------GTTTATTGCT-TTGCTGTGAACTC-CAAC------------GGAAGA-------- | |
| dp5 | 2:10611001-10611302 - | TTGGAGGCGC------ATGCGTC---GTATCCTGCGCCGTGGAA-AGCTCA-----------------------TGTTCTG---AGA----------GCCAGGGGCAGCGGACGGAGACGC----------------C-------T---------CAGA-CATTCCAGGCT---------------GAATACCGACAAAA-T--------------------------------------------------------------------------AACGG-TGTATGCAGAGAGCAGCAGAGCAGAGCAGCAAAGCAGCAAAGCAGAGCCG-----AGCAGC-AGCAGCAGCAGT-------------G-------------GCAGA-------------GC-----CAGCGCC-----AGTGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGATACAACGCACGTTG-CTGCCTCG---TAGTCGTTGT------CTG----TCTGTCGTTG-----------------------------------------------------------------TCC------TTTGGTG----------GTG-CTGCCAGA-G-------------------------A-------- | |
| droPer2 | scaffold_0:9868609-9868902 + | TTGGAGGCGC------ATGCGTC---GTATCCTGCGCCGTGGAA-AGCTCACA---------------------CT--TTG---CGG----------GCCAGGGGCTGCGGACGGAGACGC----------------C-------T---------CAGA-CATTCCAGGCT---------------GAATACCGACAAAA-T--------------------------------------------------------------------------AACGG-TGTATGCAGAGAGCAGCA--GCAGAGCAGCAAAGCAGCAAAGCAGAGCCG-----AGCAGC-AG-------------------CAGCG-------------GCAGA-------------GC-----CAGCGCC-----AGTGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGATACAACGCACGTTG-CTGCCTCG---TAGTCGTTGT------CTG----TCTGTCGTTG-----------------------------------------------------------------TCC------TTTGGTG----------GTG-CTGCCAGA-G-------------------------A-------- | |
| droWil2 | scf2_1100000004943:2557813-2558041 + | ---------CAGGCGCATGCGTT---GTATCCTGCGCCGTGGAA-AGCTCA-----------------------TT-C----------GGTCGGTCCCCTGCT---------------CGCTTGGTTGGACGGGCAGCGTATC--A---------TCAG-CATTCCAGGCT---------------GAATACCGACAAAA-T--------------------------------------------------------------------------AACGG-TATAC------------------------------------------CAGGCGA---------------------------------GT-------------A-GC-------------GC-----TT--ACC-----ATAGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACACAACGCACGCTTGCCTACT-T-GCTGTCCGTTGGC--------GTTGTCTGGCCTCT------------------------------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12822:854196-854472 + | ATGGAGGCGC------ATGCGTC---ATGTCCTGCGCCGTGGAA-AACGCA-----------------------CGGC----------AGACGTTTAGCTAC--------------------------------GAGC-------T-----------GG-AATTCCAGGCT---------------GAATACCGACAAAA-AAG-----------------------------------------------------------------------TAACGG-TA-------ACGG------------------------------------------------------------C-----AG--CAGCGC-------------G-GC-----G--------C-----GA--A-C-----AGCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACACAACGCACGCTG-CCAA-------TTGCTGTCGTT--------GTCGTCGGCCGCCCAAAA-----AA-GCAAAAGAAAACTGAAAATTCG--------A----GGA----------AAAACTTGCTGTATTTT--------------G----TGCAAAA--AATTAAAC------------GAAAAA-------- | |
| droMoj3 | scaffold_6540:33519655-33519958 + | GCCACAG-GCAGGCGCATGCGTC---ATGTCCTGCGCCGTGGAA-AGCTC------------------------CG-----AA--------------------------------AGA-----------GACCTGAGC-------T-----------GG-AATTCCAGCCT---------------GAATACCGACAAAAAC-ACACACACACAGCCACAGCCGCAGCAGCAGCAGCAGCAGCGACAGCAGCAGCAGCAGCAGAAGAAATGAAAATAACGG-TATAG--------------------------------------------------------------------C-----AG--CAGCTA-------------C-GT-------------GC-----GA--G-C-----AGCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACACAACGCACGCTG-CCAACT---G-CTGCTGTCGTT--------GC------TCACTTTGCA-----A----------------------------------------AATCCATAAAAT---------------------------------------AA--------------AATAGAAAAGAAAA-------- | |
| droGri2 | scaffold_15074:2938652-2938917 - | GTGGAGGCGC------ATGCGTCA--TAATCCTGCGCCGTGGAA-AGCTTA-----------------------CGGCTTGATACGGGAGAC---TCGCGAC-----------------------------GTGCTGC-------T----------TGG-AATTCCAGGCT---------------GAATACCGACAAAA-T--------------------------------------------------------------------------AACGG-TATAC--------------------------------------------------------------------A-----AG--CAGCGT-------------T-GG-------------GC-----CA--G-C-----AGCGCCAGACAGAAGTTACCTTTGCTCCTGCCCGGACGACACAACGCACGCTG-CCATTT---G-CTGTCGTTGTCGTTGTCTGGCC-TCGGCCTCCCACAC-----AC--------------------------ACACA--------ACTACACAAAAGAAAAT-----------------------------T-CT--------------------AGAAAAAAAAA-------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 11:41 PM