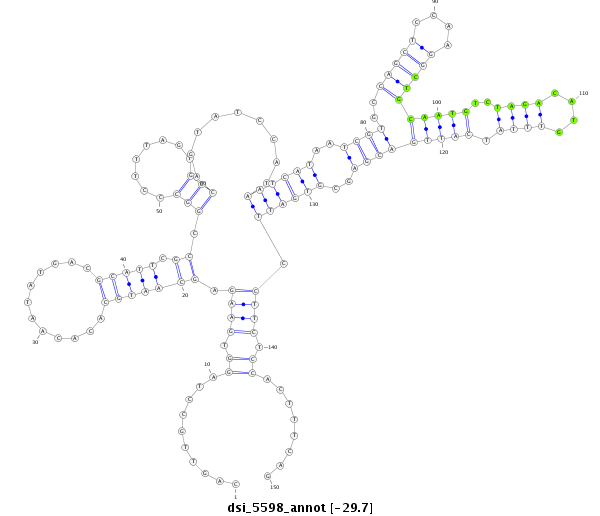

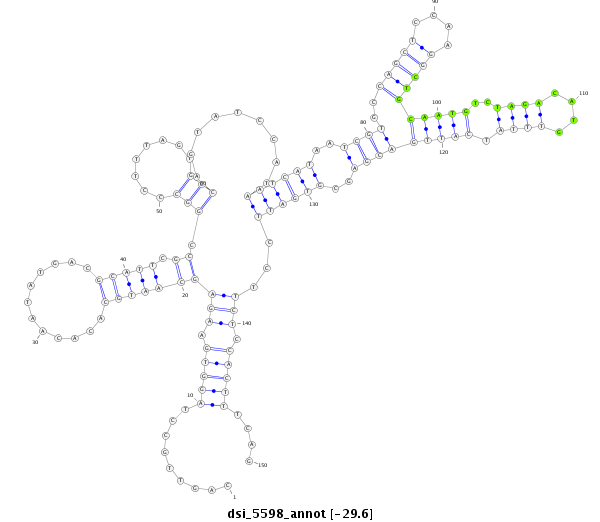
ID:dsi_5598 |
Coordinate:2r:5880047-5880196 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
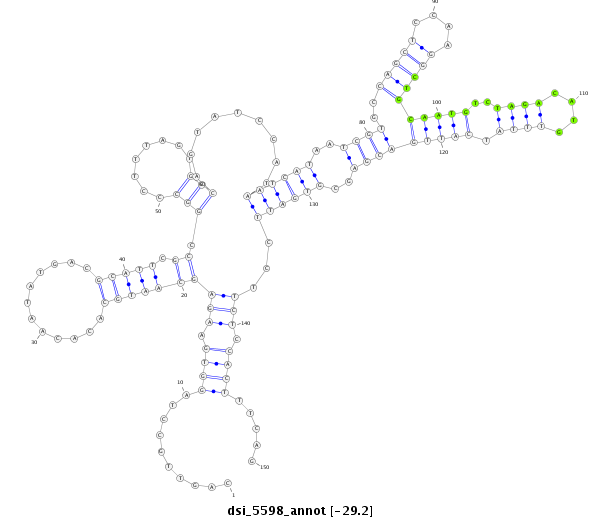
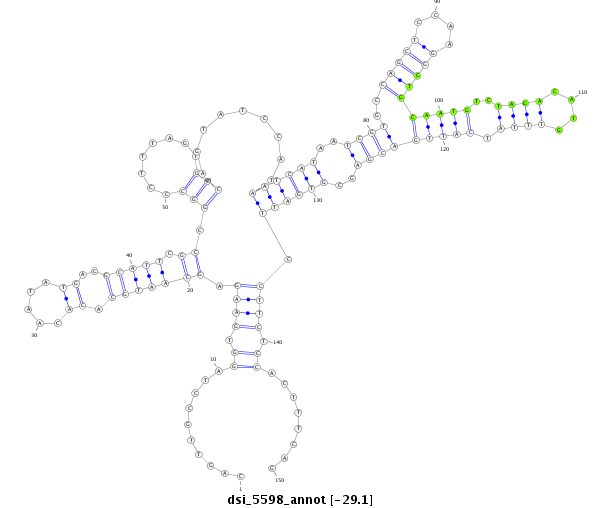
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -29.6 | -29.2 | -29.1 |
 |
 |
 |
exon [2r_5879893_5880046_-]; CDS [2r_5879893_5880046_-]; intron [2r_5880047_5882150_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GCCTGGGCTTTTTGGTTTCGGAGTCAACAACTGATATACCCGTAGACGTACAGTTGCCTAGGTGAAGAGCAATGCACACAATATGACGCATTCGCCGGCCCTTTAGTGCCAGTATCCATAATCATAATCGTGCCAGCTCCAAGGCTGCAATGTCTAGACATGTTTATCATTGACGAGCGTGATTCCTTCTCCACTTTCAGTTAACGATGGCCGGACAGCTACGGTGCCGCAATACCGGGGACTGAGCAGC **************************************************..........((.((((.(((((((............))))).)).(((........))).........((((((..((((..(((((....)))))(((((..((((....)))).)))))))))..)))))).)))).))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR902008 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
O001 Testis |
SRR618934 dsim w501 ovaries |
O002 Head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................TAACGATGGCCGGACAGCTACGGTGC....................... | 26 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................CAGTTAACGATGGCCGGAC.................................. | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................CTGCAATGTCTAGACATG........................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................TCAACCGGTAGACGTACAGTT................................................................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................ACAGTTGCCTAGGTGCAGA...................................................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CGCAGTTAACGATGGCCGGAC.................................. | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CGCAGTTAACGATGGCCGGA................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TTAACGATGGCCGGACAGCTACG........................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................GCTGCAATGTCTAGACATG........................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CCGGACAGCTACGGTGCCGCAATACCGG............ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TACGGTGCCGCAATACCGGGGACTGAGC... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CGCAATGCCGGGGACTGAGC... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....GGGCTTTTTGGTTTCGGAGTC................................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AACGATGGCCGGACAGCTACGGTGC....................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TTCGCAGTTAACGATGGCCGGACAGCTAC............................ | 29 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................CGCAATACCGGGGACTGAGC... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................GATAACCAGGGCCGGACAGCT.............................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................ATAGGCCCGTAGACGTA........................................................................................................................................................................................................ | 17 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................AGTTAAGGTGCCGGAATAC............... | 19 | 3 | 17 | 0.29 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 1 |
| .................................................................................................................................................................................................................................GCCGAAATAGCGGGGAC........ | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................CGAGCGTGATTGTTTTTCC.......................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TCTAGACATGACTATGATT............................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................ATACCGCGGACTGACC... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CGGACCCGAAAAACCAAAGCCTCAGTTGTTGACTATATGGGCATCTGCATGTCAACGGATCCACTTCTCGTTACGTGTGTTATACTGCGTAAGCGGCCGGGAAATCACGGTCATAGGTATTAGTATTAGCACGGTCGAGGTTCCGACGTTACAGATCTGTACAAATAGTAACTGCTCGCACTAAGGAAGAGGTGAAAGTCAATTGCTACCGGCCTGTCGATGCCACGGCGTTATGGCCCCTGACTCGTCG
**************************************************..........((.((((.(((((((............))))).)).(((........))).........((((((..((((..(((((....)))))(((((..((((....)))).)))))))))..)))))).)))).))........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR618934 dsim w501 ovaries |
M053 female body |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
M023 head |
O002 Head |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR902008 ovaries |
SRR1275483 Male prepupae |
SRR1275487 Male larvae |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................GCAGAGGTGTAAGTCAAT............................................... | 18 | 2 | 3 | 2.00 | 6 | 4 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................GATCCAATTCTAGTTACGT.............................................................................................................................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GGCAGAGGTGTAAGTCAAT............................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................GCAGAGGTGTAAGTCAATG.............................................. | 19 | 3 | 20 | 0.90 | 18 | 6 | 0 | 0 | 6 | 0 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................AGAGGTGAAAGTCAA................................................ | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................CCTCAGTTGTTGACCAAA..................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................AGAGGTGTAAGTCAAT............................................... | 16 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................AGGATCAACTTCTCGTTA................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TGCAGAGGTGTAAGTCAAT............................................... | 19 | 3 | 20 | 0.15 | 3 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................GCAGAGGTGTAAGTCAA................................................ | 17 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| ...............................................................................................................................................................................................................ACCGTTCTGTCGATGCCACC....................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GGCAGAGGTGTAAGTCAA................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................TGCACTATATCGGCATCTG........................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................GTTATGGCCCCTGGCG..... | 16 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................TAAGGGCCAGGTGAAAGTCAA................................................ | 21 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GGGAAGAGGTGAATCTCAA................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................AGCTGCCGGGAAATCC............................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................CTATGGGCCTGTCGAT............................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 01:48 AM