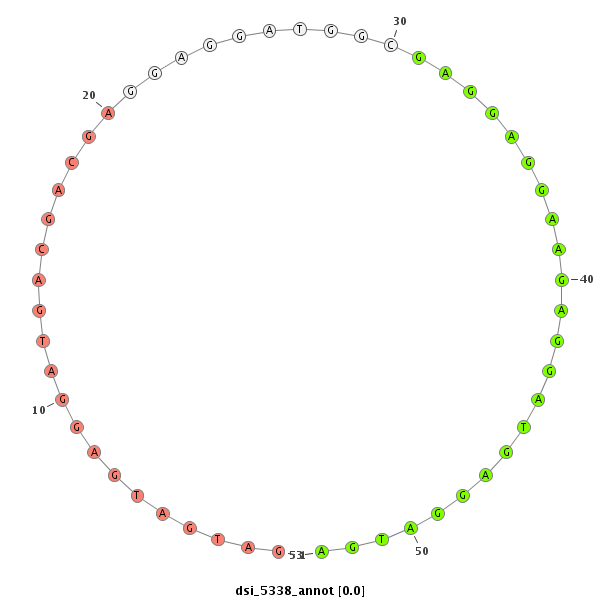
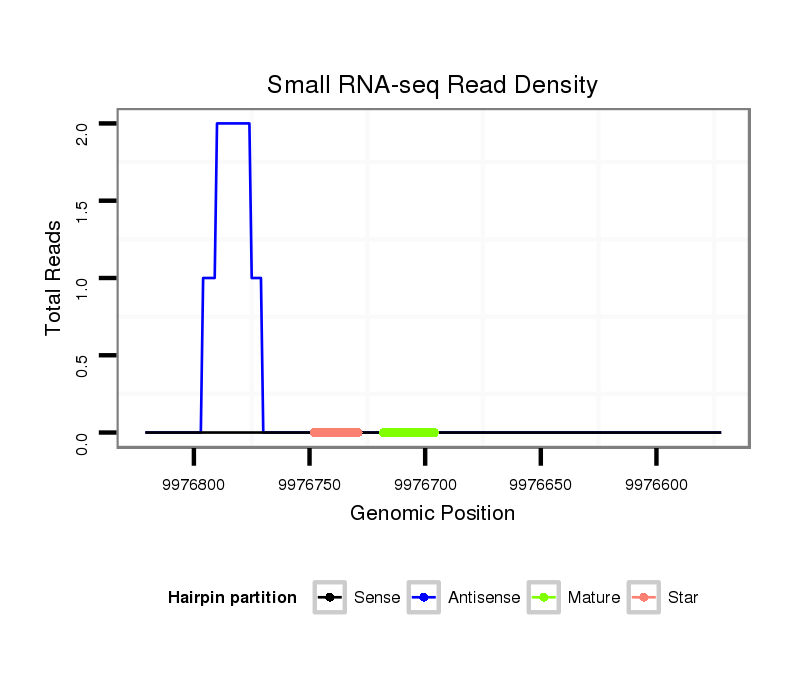
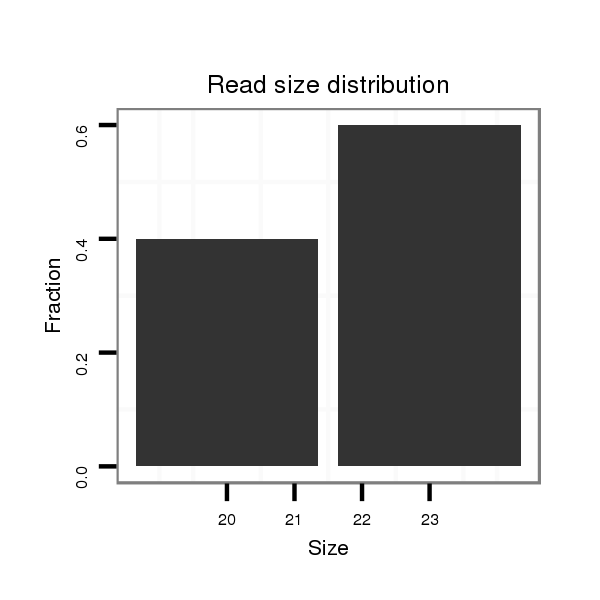
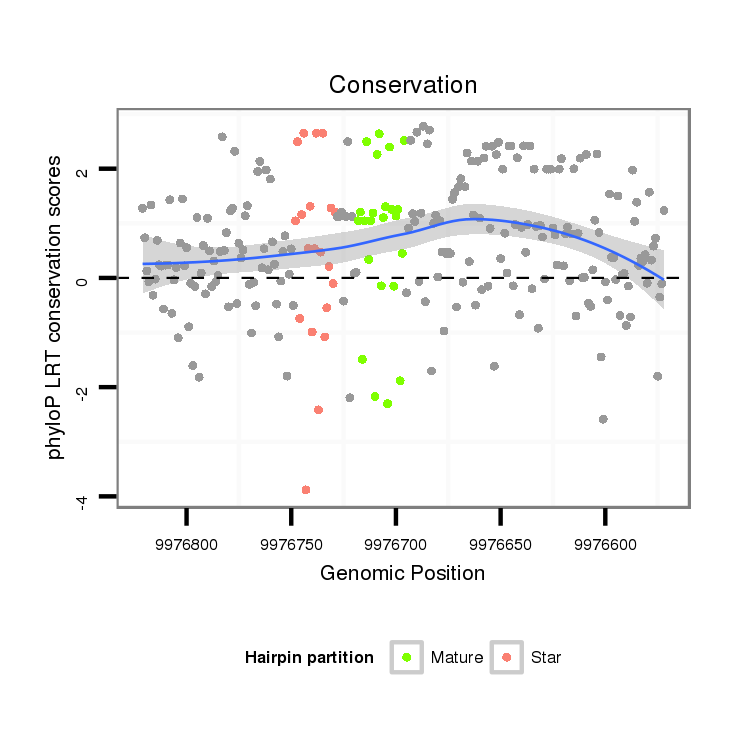
| droSim2 |
x:9976572-9976821 - |
dsi_5338 |
CAACAATAAC---------GGTAACACCAAG-----------------TCACGTGA---------TCGCGCCGATCATCATCTGCAG-----------------------------------------T------------CGCAGCAGC------------------------------AGGA---GC---------------ACGACG---ATGATGAGGATGA---CGACGAGGAGGATGGCGAGGAG---GAAGAGGATGAGGATGACGAGGAG---------C---AGCA------------------------------------------CCAG------G---AGCAGCAGCAGC---------------------------AGCA---GCAGCAGCATCAGGGCAATGG------GCA-TGATCTCACGCCCACGGACCTGCGACATC-----TGATGCCG---GCGAGCA------AC----------------------A--------------------------------------------ATATG---CAG--------------------------------------------------------------------------------------CAACAGC---------ATACG |
| droSec2 |
scaffold_15:1216025-1216262 - |
|
CAACACTAAC---------GGTAACACCAAG-----------------TCACGTGA---------TCGCGCCGATCATCATCTGCAG-----------------------------------------T------------CGCAGCAGC------------------------------AGGA---GC---------------ACGACG---ATGATGAGGATGA---CGACGAGGAGGATGGCGAGGAG---GAAGAGGATGAGGATGACGAGGAG---------C---AGCA------------------------------------------CCAG------G------------------------------------------AGCA---GCAGCAGCATCAGGGCAATGG------GCA-TGATCTCACGCCCACGGACCTGCGACATC-----TGATGCCG---GCGAGCA------AC----------------------A--------------------------------------------ATATG---CAG--------------------------------------------------------------------------------------CAACAGC---------ATACG |
| dm3 |
chrX:10511663-10511906 - |
|
CAACAATAAC---------GGTAACACCAAG-----------------TCACGTGA---------TCGCGCCGATCATCATCTGCAG-----------------------------------------T------------CGCAGCAGCAGC---------------------------AAGA---GC---------------ACGACG---ATGACGAGGATGA---CGACGAGGAGGAGGGCGAGGAG---GACGAGGATGAGGATGACGAGGAG---------C---AGCA------------------------------------------CCAG------G------------------------------------------AGCAGCAGCAGCAACATCAGGGCAATGG------GCA-TGATCTCACGCCCACGGACCTGCGACATG-----TGATGCCG---GCTAGCA------AC----------------------A--------------------------------------------ACTTG---CAG--------------------------------------------------------------------------------------CAACAGC---------ATACG |
| droEre2 |
scaffold_4690:4616163-4616415 + |
|
CAACAATAAC---------GGCAACACCAAG-----------------TCGCGGGA---------TCGCGGCGATCATCATCTGCAG-----------------------------------------T------------CGCAGCAGCAGCAAT---------CGCAGCAAGCGCAGCAGGC---GC---------------AGGACG---ATGACGACGAGGA---CGACGAGGAGGATGGCGAGGAG---GACGACGAGGAGGACGACGAGGAG---------C---AGCA------------------------------------------C------------------------------------------------------------CACCAGCAGCAGGGCAATGG------GCA-TGATCTCACGCCCACGGACCTCCGACATG-----TGATGCCG---GCGAGCA-------------------------GCAACA--------------------------------------------ACATG---CAGCA--------------------------------------------------------------------------------------ACAGC---------ATCCG |
| droYak3 |
X:19080684-19080951 - |
|
CAACAATAAC---------GGCAGCACCAAG-----------------TCACGAGA---------TCGCGCCGATCATCATCTGCAG-----------------------------------------T------------CACAGCAGCAGCAACAGCAGCAGATGCAGCAAGCGCA---------AC------AGCAGGCACAAGACG---ATGACGAGGAGGA---CGACGAGGAGGAGGGCGAGGAG---GACGATGATGAGGATGACGAGGAG---------C---AGCA------------------------------------------C------------------------------------------------------CA---TCAGCAGCAACAGGGCAATGG------GCA-TGATCTCACGCCCACGGACCTGCGACATG-----TGTTGCCT---GCGAGCA------AC----------------------A--------------------------------------------ACATG---CAGCAG-----------------------------------------------------------------------------------CAACAGC---------ATACG |
| droEug1 |
scf7180000409538:186066-186307 + |
|
CAA---TAAC---------GGCAACAGCAAG-----------------TCTCGAGA---------TCGCGACGATC---ACCTGCAG-----------------------------------------G------------CACAACAAC------------------------------AATC---GC---------------AAGATGATGAGGATGAGGAGGACGAC---T------A---TGATGAG---GATGAGGAAGATGAGGAGGAGGAG---------C---AGCA------------------------------------------G------CATCGTCACCAGCAACAGC---------------------------AGCA---GCAGCAGCAACAGGGTAATGG------GCA-TGACCTTACGCCCACGGACCTGCGGCATG-----TGCTGCCG---CCTAGCA------ATCACGATC---------------------------------------------------------------------AG--------------------------------------------------------------------------------------CAACAGCA--------ACACG |
| droBia1 |
scf7180000302126:3411981-3412255 - |
|
TAACAAT------------GGCAACAGCAAG-----------------TCGAGGGA---------TCGCGACGATC---ACCTGCAG-----------------------------------------GCACAGCAGCAGTC------------------------GCAGCAGTCGCAGCAGCC---GC---------------AGGACGACGAGGACGAGGACGAGGAC---T------A---CGACGAG---GACGAGGAGGAGGACGAGGAGGAG---------C---AGCA------------------------------------------CCAG------CAG------CAGCAGC---------------------------AGCA---GCAGCATCAGCAGGGCAACGG------GCAGTGACCTCACGCCCACGGACCTGCGACATG-----TGCTGCCC---AGCAGCA------ACCACGATCAGCA---GCAGCAGCAACATC------------------------------------------AG---CAGCAGCAG--------------------------------------------------------------------------------------C---------AGCAG |
| droTak1 |
scf7180000415394:329292-329535 - |
|
CAACAATAAC---------GGCAACGGCAAG-----------------TCGCGCGA---------TCGCGACGATC---ATCTGCAG-----------------------------------------GCGCAGCAGCAATC------------------------GCAG------CAGCAGCA---GC---------------AGGAAGATGAAGATGAGGACGAGGAT---T------A---CGACGAG---GATGAGGAGGAGGACGAGGAGGAG---------C---A------------------------------------------------------------------------------------------------------------------GCAGGGAAATGG------GCA-TGACCTCACGCCCACGGACCTGCGACATG-----TGCTGCCC---TCCAGCA------ACCACGAGCAGCA---GCAGCAACAGCAAC------------------------------------------AGCAGCAACAGCAG--------------------------------------------------------------------------------------C---------AACAC |
| droEle1 |
scf7180000491087:107855-108113 + |
|
CAATAATAAC---------AACAACAATAAC-----------------T----------------------------------ATGG-----------------------------------------CCGCAGCAGCAGTC------------------------GGAG------CAGCAGCA---GCTCACAAAGCAGCC--TGGACGAAGAGGAGGACGACGAGGAG---G------A---CGAAGAG---GACGAGGAGGAGGATCAGGAGGAGGCAGTGTCCC---AGCA------------------------------------------CCAG------C------------------------------------------AGCA---GCAGCAGCAACA----------------------------------------------AC-----T--GGCGGCCGAGCAACA------ACAACGGGCAGCT---GCAGCAGCGGCCGTCGTGGCAGCGGCGGTCAGTGCGC-TGCACAACG---GCAACA------AGCAGCAG--------------------------------------------------------------------------------------C---------AGCAG |
| droRho1 |
scf7180000780277:35867-36140 - |
|
CAACAATAAC---------GGCAGCAGCAAG-----------------TCACGAGA---------TCGTGACGATC---ACCTGCAG-----------------------------------------GCACAGCAGCAACA---------------------------GCAATCGCAGCAGCC---GC---------------AGGACGAAGAGGACGAGGAGGACGAC---T------A---CGACGAG---GATGAGGAGGAGGACGAGGAGGAG---------C---AGCA------------------------------------------TCAG------CAG------CATCAGC---------------------------AGCA---TCGCCAGCAGCAGGGCAACGG------TCA-TGATCTAACACCCACAGACCTGCGACATG-----TCCTGCCG---CCCAGCC------ATCACGATCAGCA---GCAGCATCAGCAAC------------------------------------------AG---CAGCAGCAA--------------------------------------------------------------------------------------C---------ATTTG |
| droFic1 |
scf7180000454050:471713-472001 - |
|
CAACAATAAT---------GGCAACAGCAAG-----------------ACGCGAGA---------TCGCGACGATC---ACCTGCAG-----------------------------------------GTGCAGCAGCAATC------------------------GCAG------CAGCAGGC---GC---------------AGGACGAAGAGGATGAGGAGGATGAC---T------A---CGATGAG---GATGAGGAGGAGGACGAGGAGGAG---------C---ACCA------------------------------------------ACAG------CAG------CAGCAGCAACAGCAGCAGGTAAACCGCCACCAGCAGCA---GCAGCAGCAGCAGGGCAACGG------GCA-TGATCTCACGCCCACCGACCTGCGACATG-----TGTTGCCC---GCCAGTC------ATCACGATCAGCA---ACAGCAG------------------------------------------------------CAACATCAG--------------------------------------------------------------------------------------C---------AGCAG |
| droKik1 |
scf7180000302698:593192-593441 + |
|
CAATAACAAT---------GGCAACAGCAAG-----------------CGA---GATCAGCGGGAACGTGAAGATC---ACTTGCAA-----------------------------------------T------------CGCAGCAGC------------------------------AGCAGGCGC---------------CGCCAG---AGGAAGAGGATGAAGAC---T------A---CGACGAG---GAGGAGGAAGATGATGAAGAGGAA---------GATCACCA------------------------------------------CCAC------C------------------------------------------ATCA---GCAGCAGCAGCAGGGCAATGG------TCA-TGATCTGGGACCCACAGATCTCAGGCATG-----TCCTGCCC------------------CATGATCCACA---GCAGCAGCAACAGT------------------------------------------TG---CAGCAACAA--------------------------------------------------------------------------------------C---------AACAG |
| droAna3 |
scaffold_13117:4521667-4521898 - |
dan_1383 |
CAACAACAAC---------AACAATAACAAG-----------------TTAAGATT---------AAGCGAAGA------------A-----------------------------------------C------------CGCAGCAGC------------------------------AGCA---GC---------------ATCATGACGACGATGATGAAGATGAT---T------A---TGATGATGATGAGGAGGACGAGGATGATGAGGAT---------C---ATCA------------------------------------------TCAT------C------------------------------------------ATCA---TCATCAGGATCAGGGTAATGG------ACA-CGATCTAACACCCGAAGATCTGCGGCACA-----TGATAC---------------------ACGATCCCCA---GCAGCAGCAGCAGC------------------------------------------AG---CAGCAACAG--------------------------------------------------------------------------------------T---------CGCAG |
| droBip1 |
scf7180000396022:3538-3769 - |
|
CAACAATAAC---------AGTAGCCACAAG-----------------CTAAGATT---------AAGCGAAGATCC------GCAG-----------------------------------------CAGCAGCAACAGTC------------------------GCAG------CAGCAGCA---GC---------------AGCATGAAGACGATGATGAAGACGAT---T------A---CGATGATGATGAAGAGGATGAGGAAGATGAGGAT---------C---ATCA---------------------------------------------------------------------------------------------------------TCAGGATCAGGGTAATGG------ACA-CGATCTAACGCCCACAGATCTGCGGCACA-----TGATAC---------------------ACGATCCGCA---GCAGCAGCAACAGT------------------------------------------CG---C------AG--------------------------------------------------------------------------------------C---------AGCAG |
| dp5 |
XL_group1e:5881827-5882097 - |
dps_2709 |
CAACATTAAC---------AACAACAGCAAG-----------------GGGCGAGAGCTACGAGAACTGCACGATCC---TCAGCAG-----------------------------------------CAGC---------AGCAGCAAC------------------------------AGCA---AC------ATC------ATCCAGAAGATGAGGAGGAAGACGAC---T------A---CGACGAGGATGATGAGGACGATGATGAAGAGGAGCCA------C---AACA------------------------------------------CCAG------C------------------------------------------AGCA---GCAGCAACAACATGGCAATGGACTTGGCCA-TGACCTCACGCCCACAGATCTGCGTCACG-----TCCTGCCA---TCCAGCACCACCCATCACGATCATCT---CCAC---------------------------------------------------------CAGCAG-----------------------------------------------------------------------------------CAACA------------GCAG |
| droPer2 |
scaffold_0:5813869-5814113 - |
|
CTGAGATGGAG--------GAGACGACCATG-----------------TCG--------------CCGCCGCAATCATCATCGG-AG-----------------------------------------C------------AGCAGCAGC------------------------------AGCA---GC---------------ACCACC---ACCACCACCATCA---GCACCAGGACGAGGACCCGCAG---CG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCCCGAAATGAAGAATCTAGCCA------------------CACTTTGTTCAGCAGCAGCAGCGGCCGC------------------------------------------------TGCTGTGGCAGCAGGCCACAAGGATGAAGATGACCTCAGTACAGATCAATACAGTGATGAGGACGATGCTATGGGC------------GATGAGA---------ATACG |
| droWil2 |
scf2_1100000004515:3771087-3771348 - |
|
CACAAGCAGCAACAACAACAGCAGCAGCAAG-----------------TCAAGAGA---------TCATCTGGAAG---AGCAGCAA-----------------------------------------T------------CACAGCAGC------------------------------CAGC---AG---------------AAGACG---ATGATGAAGACGA---T---T------A---CGATGACGATGATGACGATGAGGACGAAGAAGAA---------C---ACCA------------------------------------------GCAA---CATCAA---------CACCAACACCAACAG---------CAGCAGCAGCA---ACAACAACAACAAGACAATGG------ACA-TGATCAAACGCCCACAGACCTGCGCCATG-----TTCTGCCG---C------------------------------AGCATCATCAAC------------------------------------------AG---CAGCAACAA--------------------------------------------------------------------------------------C---------ATCAG |
| droVir3 |
scaffold_12970:7508781-7509002 + |
|
CCGCACTGCC---------CGCCGATTTGCG-----------------GCACGGAATCAGC----TCG-CACGAT---CA------G-----------------------------------------A------------GCCAGGAGC------------------------------CGGC---GG---------------ACTACG---ATGACGAGGATGA---GGACGAGGAAGACGATGACGAG---GATGAGGATGCGGC-------------------C---AGCAGTCATCTGGACAGCCGCACGGCGCTAACATCAACGCATCTGAGCAG---CAGC-------------------------------------------------------------------------------------------------------------------GCCGCC-----------------------------------------GCCGAG---AGCAGCGGC-----TTGCACAT--------------GAGCG---CAGTGGCCG--------------------------------------------------------------------------------------C---------GGCCG |
| droMoj3 |
scaffold_6500:2415650-2415866 + |
|
CAGCAGCAAC---------AGCAGTTCCAAG-----------------CACCATCG---------GCGCGACAAGCATCAC-AGCAA-----------------------------------------A------------AGCAC---------------------------------CAGCA---GC---------------ACT--------GAGATCAATCA---TACCG------------------------AATTGGATAACAAGCAGCAA---------C---AGCA------------------------------------------ACAG------CAA------CAACAGC---TGCAACAG---------CAGC---AGCA---G------------------------------------------------------------------------------------------------CAGCA---GCAGCAGCAACAAC---AGCAGCAG------TTGCACACGCTCAACACCAGCGAGGAG---GAG--------------------------------------------------------------------------------------CAACAGCAGCCCTCCCAGTCG |
| droGri2 |
scaffold_15203:7100689-7100918 - |
|
CAACAACAAC---------GGCAACAACATCAGCTAGGCGACGACAAGCCGTGC----------------------------AACAACCGCACCAACGTCGAACAACGCCAATTAGCGGCGAGGGCGGCGGCAGCAGCAGTC------------------------GCAG------CAGCAGCA---CC---------------A---------------------------------------------------------------------GCAG---------C---AGCA------------------------------------------CTTGCAG---CAG------CAGCAAC------------------------------G---ACAGCAGCATCAG--CAACAG------CAA-CAAA------------------------------------------------------------AACACCA---ACAGCAG------------------------------------------------------CAACAACAA----------------------------------------------------------CAACACCGGCAACAAATAATAACCATAAC---------ATCCG |