ID:dsi_5285 |
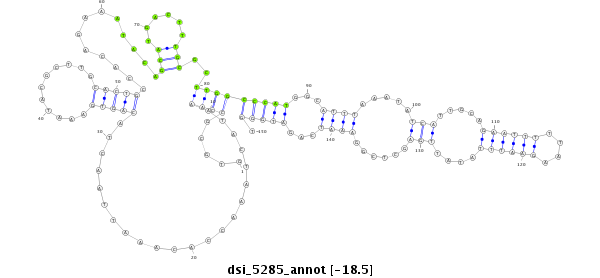
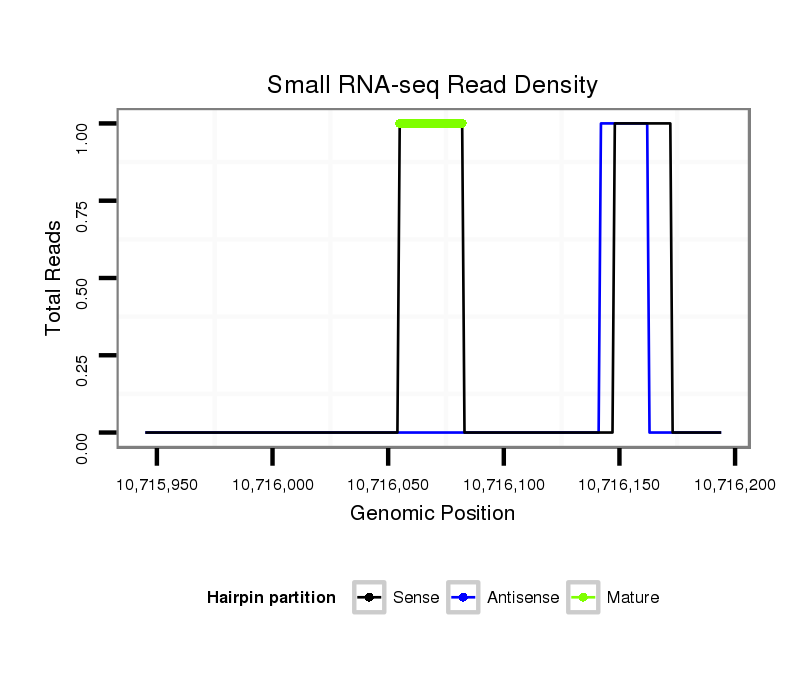
Coordinate:3r:10715995-10716144 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
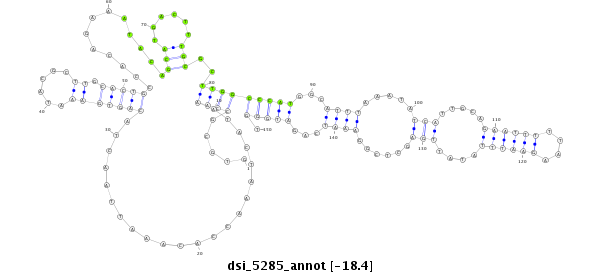
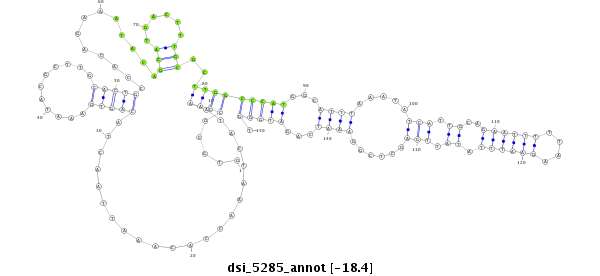
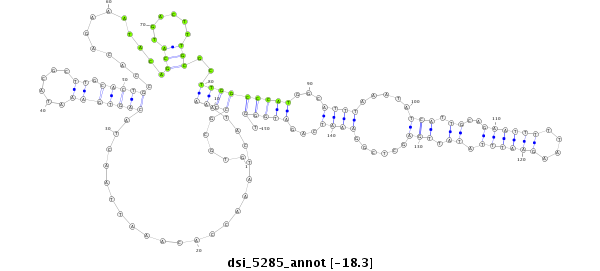
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -18.4 | -18.4 | -18.3 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCACTGGCACTCACCGAGGGCGCCCAGTTCATACAGAGCGACCCCGCGAGGTGCGCCAAATACTAAACCACAAATTAACTACAGTGAAATACGCTTGCACTGCCACAGAAATACAGCATGACTTTGCGCTTGGCCCATGGCATTTAAATATCATTGCAGAATTTTTAAGAATTTATATTGAGCTCGGAAATCAGATGGGTTCAGTGTTGTAGAACAAGAATTATAACCCTATCCTAGGTTCTTTTTATAT
**************************************************.....((((......................(((((...........))))).............(((......)))..))))(((((...((((.....(((.....((((((....))))))....)))......))))...))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
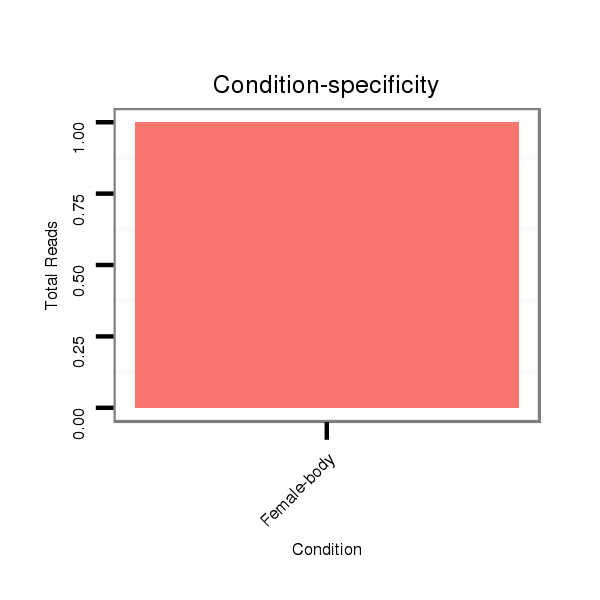
M053 female body |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................GTGTTGTAGAACAAGAATTATAACC...................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................ATACAGCATGACTTTGCGCTTGGCCCAT................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGCGATTGGCTGATGGCATTTA........................................................................................................ | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CAGTCTTGTAGAACAA................................. | 16 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................TTGCGATTGGCTGATGGCATT.......................................................................................................... | 21 | 3 | 10 | 0.30 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| ............................................................................................................................TGCGATTGGCTGATGGCATTT......................................................................................................... | 21 | 3 | 12 | 0.25 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................................TGCGACCCCGCGAGG....................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................ACCGAGCGACGCCGCG.......................................................................................................................................................................................................... | 16 | 2 | 18 | 0.11 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................ATGGGTTGAGGGTTGTA....................................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................TTGCGATTGGCTGATGGCAT........................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................CCAGTTCATACAAAG.................................................................................................................................................................................................................... | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................AGAAATTCAGCAGGACT............................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................TGTAGGAGAACAAGAAT............................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GGTGACCGTGAGTGGCTCCCGCGGGTCAAGTATGTCTCGCTGGGGCGCTCCACGCGGTTTATGATTTGGTGTTTAATTGATGTCACTTTATGCGAACGTGACGGTGTCTTTATGTCGTACTGAAACGCGAACCGGGTACCGTAAATTTATAGTAACGTCTTAAAAATTCTTAAATATAACTCGAGCCTTTAGTCTACCCAAGTCACAACATCTTGTTCTTAATATTGGGATAGGATCCAAGAAAAATATA
**************************************************.....((((......................(((((...........))))).............(((......)))..))))(((((...((((.....(((.....((((((....))))))....)))......))))...))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
M023 head |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................CCAAGTCACAACATCTTGTTC................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................AGCTCCACGCCGTTTATGA.......................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................TCTCGCTGGTGCGCGCCACT.................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................ATATTGGTCTTAATATTGG...................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................CCCGGGGATCAAGTTTGTC...................................................................................................................................................................................................................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................AACCGGGTACCATATA......................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| ..................................................CTCGCGGCTTATGATT........................................................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................CCCGGGGGTCAGGTTTGTC...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:47 AM