ID:dsi_4983 |
Coordinate:3l:8263676-8263825 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
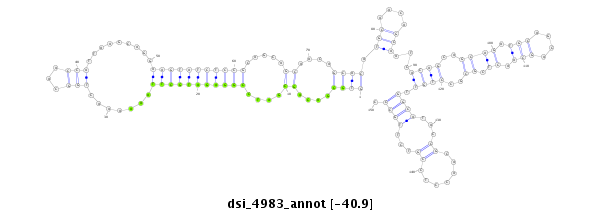
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -40.2 | -40.0 | -40.0 |
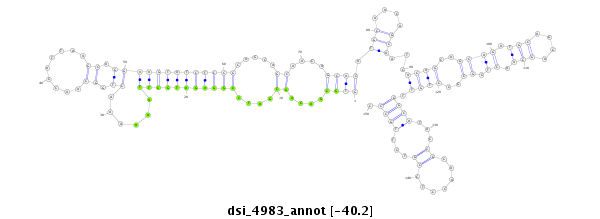 |
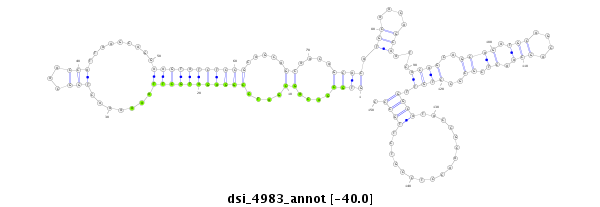 |
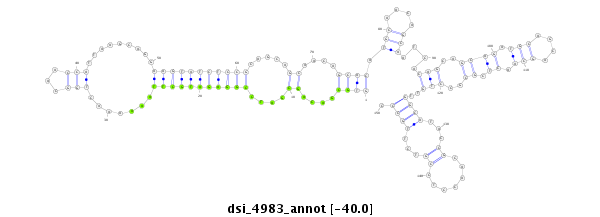 |
intergenic
| Name | Class | Family | Strand |
| TART_DV | LINE | Jockey | + |
|
GCGCTGAACGCGTCCGTCGACCGCAAGGAGGCCATCGAGAAGATGGAGTCGTGCGACAGCGATCCGGAGATGCTTAAAAAACTGGCAACCATTAAGCAGGAAGTATCTCCGCAGCAGCAACAGCACATGCAACAGCAATCACAGCAGCAGATGCAGCAGCAACTCGCGCCTGTTGGCATACCGCAACCTCCGTCTTGCCCGCCTTCGGAATCAGTCTATATCAAAAAGGAGCCCATGGAGGACTCGATGG
**************************************************((((....((....(((((((((((.......(((...))).........))))))))))).....))....)))).(((....)))...((((..((((.(((....))).)).))..)))).((((...((.......))...)))).************************************************** |
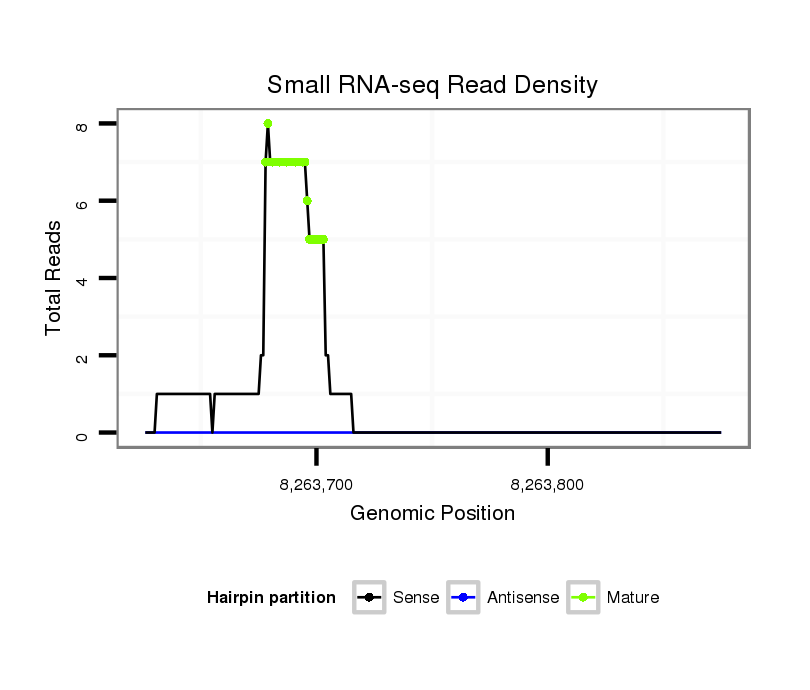
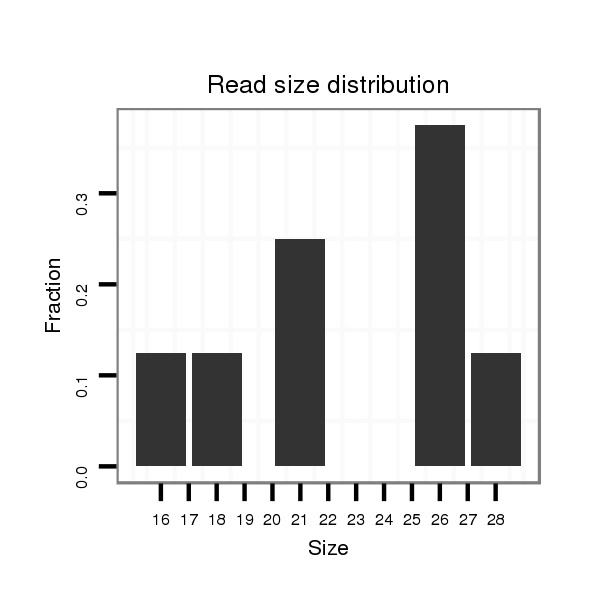
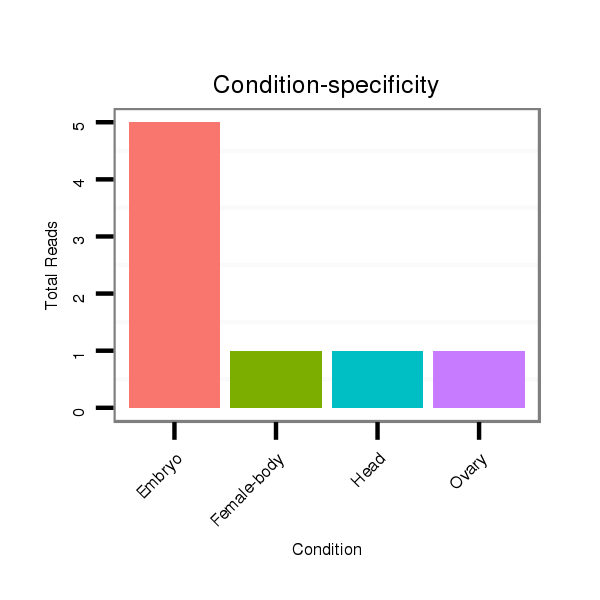
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M024 male body |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................CTTCGGTATCAGTATATATCA........................... | 21 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GCGACAGCGATCCGGAGATGCTTAAA............................................................................................................................................................................ | 26 | 0 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GCGACAGCGATCCGGAGATGCTTAAAAA.......................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTGCGACAGCGATCCGGAGAT................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....GAACGCGTCCGTCGACCGCAAGGA............................................................................................................................................................................................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................GCGACAGCGATCCGGAGA.................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................CGACAGCGATCCGGAGATGCT................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................GCCATCGAGAAGATGGAGTCGTGC.................................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................GACAGCGATCCGGAGATA.................................................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TAAAAAACTGGCAACC................................................................................................................................................................ | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................TCGGTATCAGTATATATCAGA......................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............CGTTAACCGCAAGGAGGC.......................................................................................................................................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................................GTCGTGAGACAGGGATCCG........................................................................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CTGATGACGCAGCAACTCGC................................................................................... | 20 | 3 | 11 | 0.36 | 4 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 |
| ........................................TGATGGAGTCGTGAGACAG............................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................CAATATGGAGTCGTGCGA.................................................................................................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................TGGAGTCGTGAGACA................................................................................................................................................................................................ | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................AGGAGGCCATCGAGAA................................................................................................................................................................................................................. | 16 | 0 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................AAGATGCTGTCGTGAGACAG............................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............CGCTGACCGCAAGGAGGC.......................................................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................................................CTAGGAGCCCATGAAGGAC....... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....GATCGGGTCCGTCGAC..................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................TCTTGACCGACTTCGGA......................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
CGCGACTTGCGCAGGCAGCTGGCGTTCCTCCGGTAGCTCTTCTACCTCAGCACGCTGTCGCTAGGCCTCTACGAATTTTTTGACCGTTGGTAATTCGTCCTTCATAGAGGCGTCGTCGTTGTCGTGTACGTTGTCGTTAGTGTCGTCGTCTACGTCGTCGTTGAGCGCGGACAACCGTATGGCGTTGGAGGCAGAACGGGCGGAAGCCTTAGTCAGATATAGTTTTTCCTCGGGTACCTCCTGAGCTACC
**************************************************((((....((....(((((((((((.......(((...))).........))))))))))).....))....)))).(((....)))...((((..((((.(((....))).)).))..)))).((((...((.......))...)))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M024 male body |
M023 head |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................GAAGCCATAGTCATATATAGT........................... | 21 | 2 | 1 | 8.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................AGGAAGCCATAGTCATATATA............................. | 21 | 3 | 1 | 8.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AAGCCATAGTCATATATAGTC.......................... | 21 | 3 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GGAAGCCATAGTCATATATA............................. | 20 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................TGGAAGCCATAGTCATATATA............................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAAGCCATAGTCATATATAGTC.......................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................TAGCGCTTCTACCTCATCA...................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................GTAGCGCTTCTACCTCATC....................................................................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................TATTGCGTCGGAGCCAGAACGG................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................TAGCGCTTCTACCTCATC....................................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGGGGCGTCATCGTTGTCG.............................................................................................................................. | 19 | 2 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..........................................................................................................................................................................................................GAAGCCATAGTCATATATATT........................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TCGCGTACCTCCAGAGC.... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................GGCGTTGGAGGCCGTAC..................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................TGGCGTTGTACGCAGAA...................................................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/24/2015 at 10:47 AM