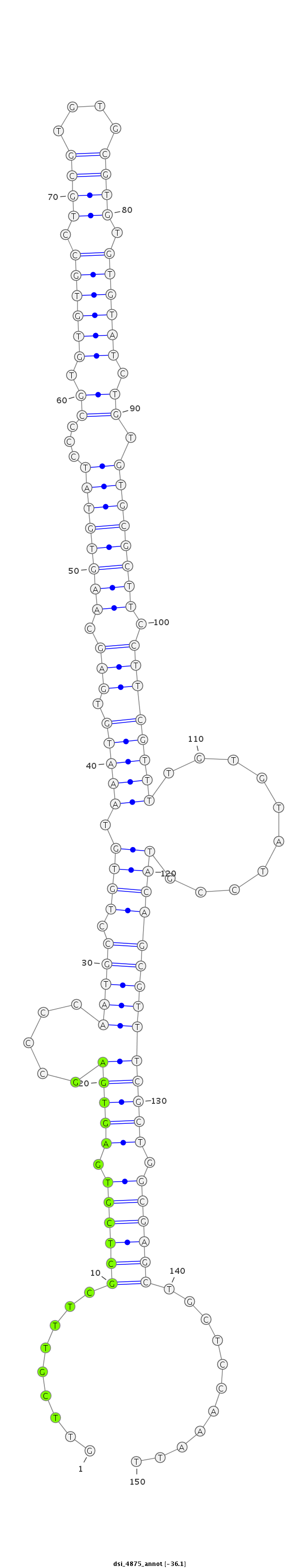
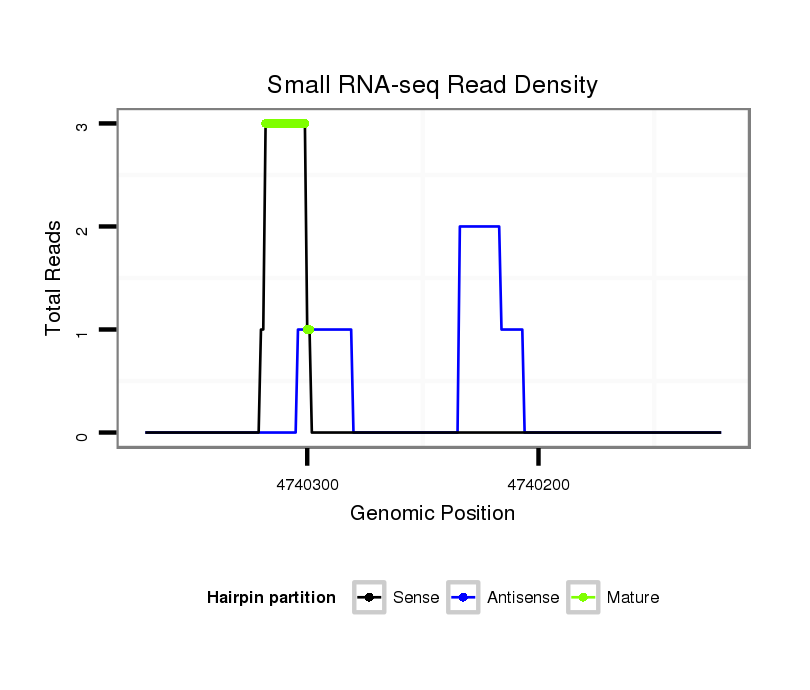
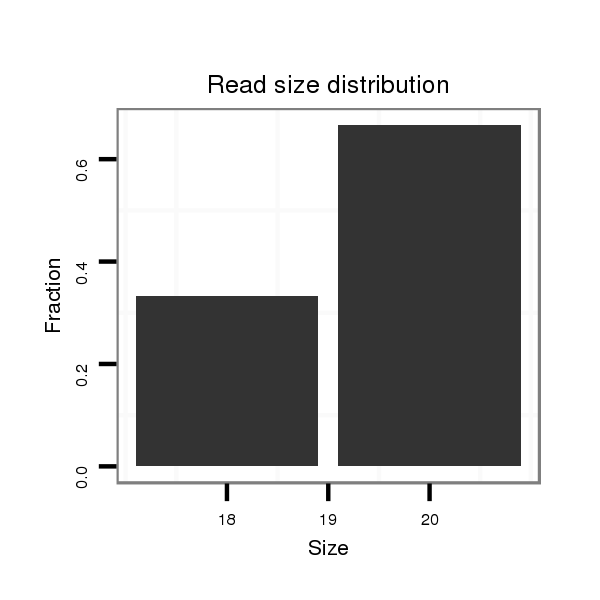
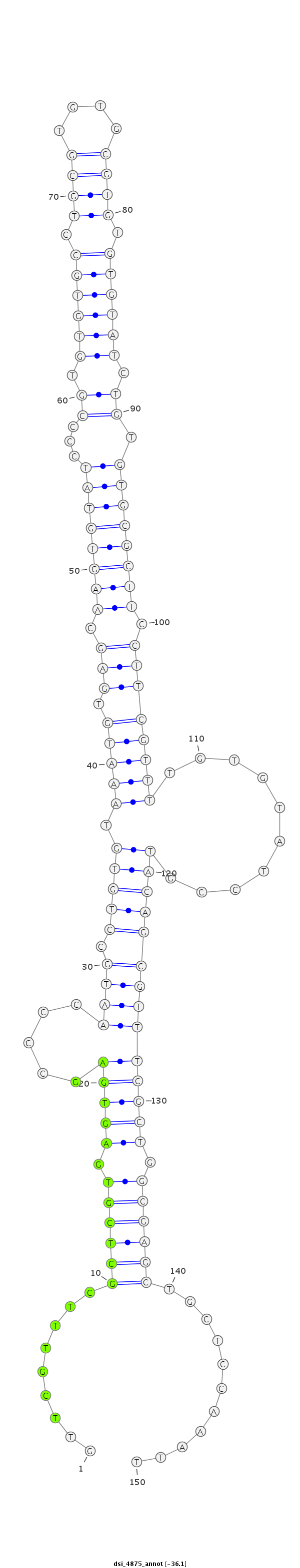
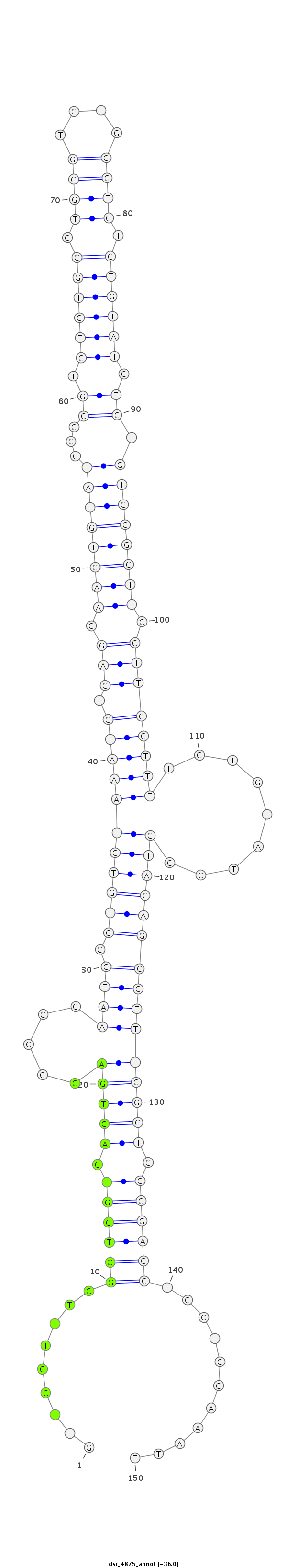
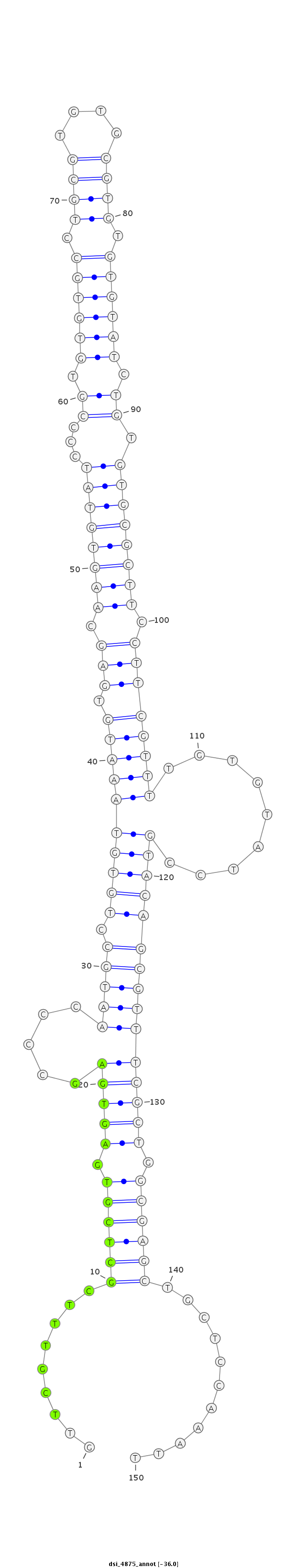
ID:dsi_4875 |
Coordinate:3r:4740171-4740320 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -36.1 | -36.0 | -36.0 |
 |
 |
 |
exon [3r_4740321_4740531_-]; five_prime_UTR [3r_4740321_4740531_-]; intron [3r_4733997_4740320_-]
| Name | Class | Family | Strand |
| Homo6 | DNA | hAT-Pegasus | - |
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTCAAAAGGCTCGTTCAAGTGGCAGTTGCTTTTTTTGGGCCAAGCCAGCGGTTCGTTTCGCTCGTGAGTGAGCCCCAATGCCTGTGTAAATGTGAGCAAGTGTATCCCCGTGTGTGCCTGCGTGTGCGTGTGTGTATCTGTGTGCGCTTCCTTCGTTTTGTGTATCCGTACAGCGTTTCGCTGGCGAGCTGCTCCAAATTTATACCCAGCATATTTTTGCACATTTAATTGAATTAAGTAAAGGGCTGAG **************************************************.........((((((.(((((.....(((((.((((.(((((.(((.((((((((...((.((((((.((((....)))).)))))).)).)))))))).))))))))..........)))))))))))))).))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................TCGTTTCGCTCGTGAGTGAG.................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTTCGTTTCGCTCGTGAGTG.................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................TCGTTTCGCTCGTGAGTG.................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TCGTTCTGTGTATCC................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................ACGCCAGCGGTTCGTCT................................................................................................................................................................................................ | 17 | 2 | 15 | 0.13 | 2 | 0 | 0 | 2 | 0 | 0 |
| ................AAGTGGCAGTTTAGTTTTTT...................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................GAATTGAGTATAGGGGTG.. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
|
CAGTTTTCCGAGCAAGTTCACCGTCAACGAAAAAAACCCGGTTCGGTCGCCAAGCAAAGCGAGCACTCACTCGGGGTTACGGACACATTTACACTCGTTCACATAGGGGCACACACGGACGCACACGCACACACATAGACACACGCGAAGGAAGCAAAACACATAGGCATGTCGCAAAGCGACCGCTCGACGAGGTTTAAATATGGGTCGTATAAAAACGTGTAAATTAACTTAATTCATTTCCCGACTC
**************************************************.........((((((.(((((.....(((((.((((.(((((.(((.((((((((...((.((((((.((((....)))).)))))).)).)))))))).))))))))..........)))))))))))))).))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O002 Head |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................AGACACACGCGAAGGAAGCAAAACACAT...................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................AGACACACGCGAAGGAAG................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................TCACTCGGGGTTACGGACACATTT................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................ATGGGCATGTTGCAAA........................................................................ | 16 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:25 AM