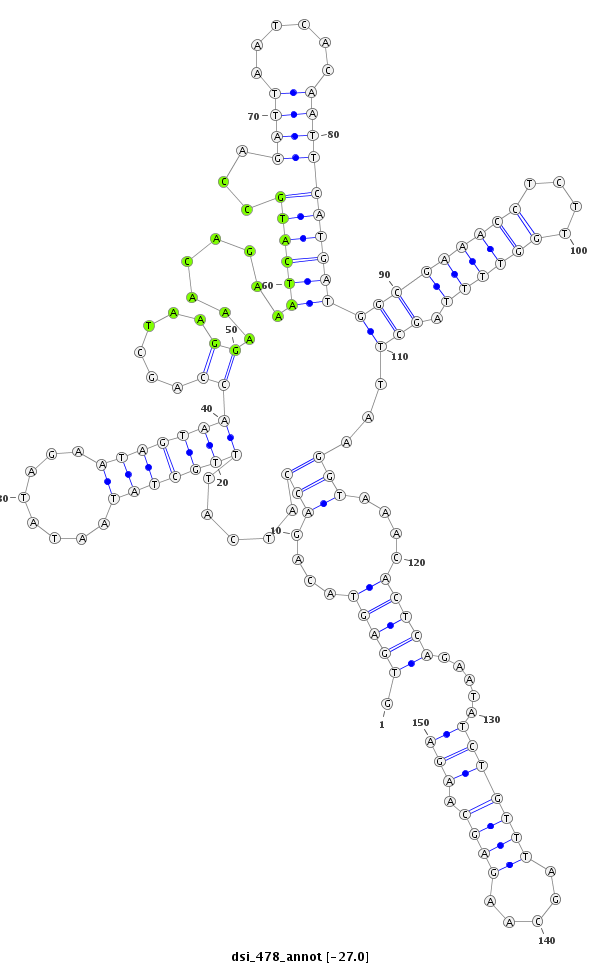
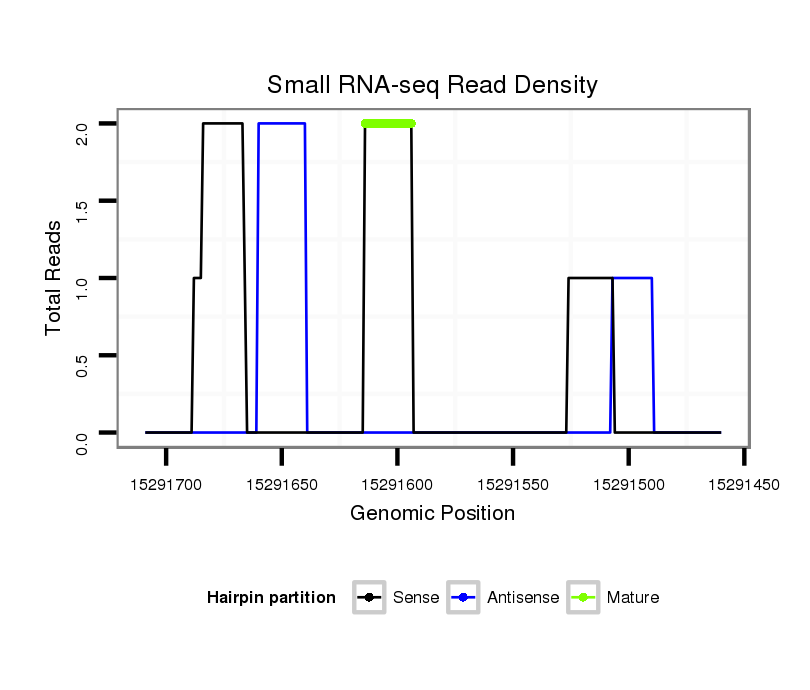
| droSim2 |
3r:15291460-15291709 - |
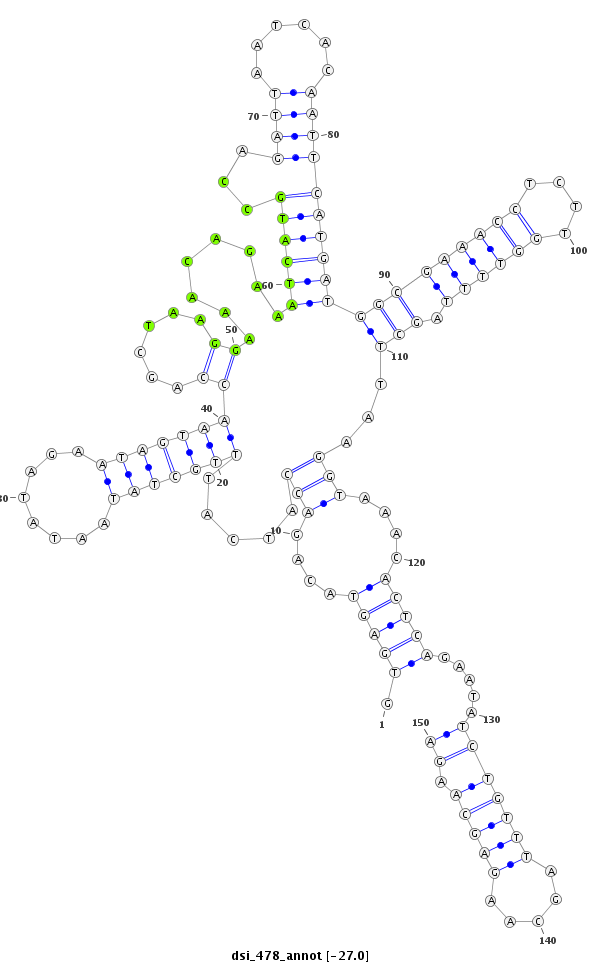
dsi_478 |
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACAGACC--------------------------ATCA-------TTTGCTATAATA----------TAGAATAGTA-----------------------------------------ACCAGCTAAGGAAACAGAAAT--------------------------------CATGCCAGATTAATCACAATTCATGA-------------------------------------------------------------------------------------------------------------------------------------TGGCGAAACCTCTTGGTTTTAGCTTAAGGTAAACACTCAGAATATCTGTTTA------------GCAA-G----------------------------------------------------------------------------AGCAAGAGACGGCC-----------AATC----------------------------ATTTTGCCGGTCCTACCCGGTTTTT-AATCGCAGC----TTTA-----T |
| droSec2 |
scaffold_0:16160805-16161049 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACAGACC--------------------------ATAA-------TTTGCTATAATA----------TAGAATAGTA-----------------------------------------ACCAGCTTAGGAAACAGAAAT--------------------------------CA-----------------------------------ACCATGGAATAGAGAA----------------------------------------------------------------------------CTCA--------------------------TGGCGAAACCTCTTGGTTTTAGCTTAAGGGAAACACTCAGAATATCTGTTTA------------CCT--G----------------------------------------------------------------------------AGCAAGAGACGACC-----------AATC----------------------------ATTTTGCCGGTCCTACCCGGTTTTT-AATCGCAGC----TTTA-----T |
| dm3 |
chr3R:5756218-5756495 + |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACAGACG--------------------------CTAA-------GTTTCTCAAATA----------TAGTATAGTA----------------------------------------AACCAGCATAGGAAACAGAAAT--------------------------------CATGCCAGATTAATCACAATTCATGATG----GTTGGCCCCAGGAATAGAGAA--------------------------------------------------------------------------ACCTAA--------------------------TGGCGAAACCTCTTGGTTTAAGCTTAAGGTAAACACTCAGAATATCTGTTTA------------CCT--G----------------------------------------------------------------------------AGTAAGA-ACGGCC-----------AATC----------------------------ATTTTGCCGGTCCTACCCGGTTTTT-AATCGCAGT----TTTA-----T |
| droEre2 |
scaffold_4770:15847182-15847556 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACAGCCC--------------------------CTAG-------TTATCCAAAGTG----------TAGTACGGTA-------------GAGTTGAGATGGCAAGGAGGTCAGTAGAACCAGCTTAAGAACCGGAACT--------------------------------CACGGAAGATGAATCAAAA-TCATGATG----ATAGGCCCCAGGAATAGAGAACTCATGTCGAAGCATCTTATTTTTAGGAATAGAGAACATATGGCATGACCTCTTGATTTTGGGAATAGAG----AACTTG--------------------------TGGCGTTACCTCTTGGTTTTAGCCCATAGTAAACAACCAGAATGTCTGTTTA------------CCT--A----------------------------------------------------------------------------GGCAAGAAACCGCC-----------GCTT----------------------------ATTTTGCCGGTCCTACCCGGTTTTT-AATAGCAAT----TTTA-----T |
| droYak3 |
3R:9802830-9803207 + |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACAGCCC--------------------------TTAA-------TTATTGACAATA----------TGCTATAGTA-------------GAGTTGAGACTGTAAAGAAGTCAGTACAACCAGCTAAAAGGCTAGAAAT--------------------------------CATGGAAGATTAATCAGGAATCATGCTG----ATAGGCCCCAGGAAACGAGAATTCATGTCAAAGCCTCTTGGTTTTAGGAATAGAGAATTTATGTCAAAACCTCGTGGTTTTAGGAATAGAG----AATTCA--------------------------TGTCAAAACCTCTTGGTTTCAGCTCAAAGTAAACACCCAGAATATCTGTTTA------------CCTATA----------------------------------------------------------------------------GGCAAGAAGCCGCC-----------GTTC----------------------------AGTTTGCCGGTCCTACCCGGTTTTT-AATCGCATT----TTGA-----T |
| droEug1 |
scf7180000409470:76104-76390 + |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACTAGCT--------------------------ACTATATAA--TTA---TTAATATGAAAAG--T---------ATCTAGATACATG-AAAATGATATGGTAAAGAAGTTGGCTTAACTAGC--------------------------------------------------------------------------------------ACAGGAAAAGAGAGAGAGTTTCAAAGAC----------------------------------------------------------ATATCTAGTCAAAAT-----------------------GT-A--TC-----TTTAACT--------------TGAGTATCTGTTTTGATGTGTACCTACCTATA----------------------------------------------------------------------------GGTAGGC---TTAC-CTTACCCCACTATT----------------------------TTTTTGCCGGTCTTACCCGGTTTTTCTTTCGCA-T----TTTA-----T |
| droBia1 |
scf7180000302402:3247006-3247283 + |
|
ACCATCCAAAACAGTATTGAGGTGCC---------------GACGA---TGCAGAAGTGAGTACATATG--------------------------AACT---------------TATATTCAAAGGAT---------ATATAGAGACACCAAAAAT--------------------------------------GGGACATTTTGGAGCAGATGGC-----CTAAGGCTTTT-----CAG-----T-------------------------CCA-AAACCAATATAGCCTTTCAAAAAC------------------------------------TACC---------------GAATTACCCAACCTGACT-----------------------TG-T--TG-----TTTACCT--------------TAAATATATATTCTGC-----GCTTGCCTAGA----------------------------------------------------------------------------GTTAAGAAACCGTC-TGTCCCCCACTA-T----------------------------TTTTCGCCGGTCGTACCCGGTTTTC-ATTCGCA-T----TTCA-----T |
| droTak1 |
scf7180000415245:366703-366993 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTGACGGCGC---AGCACAAGTGAGTACTACTTAATAACTATGGAT----------------A---------------AATAT--------T---------ATCAAGAAATAC-----------------------------------------------AAAATTCTGGAGCCTATAGTAAAATCCAAGGTTCCC-----------------------------------------AAGTACTAAGAT--------------------------------------------------------------------------ACCAAACGTAACTCGACCGGCTAACTAGAATTTTGTGT-ATCTC-----TTTAACTTTACA---------TGAATATCAGTTTAGACGTATA--AACC---A----------------------------------------------------------------------------AGTAAGAAACCATTATGTCCCCCACT-TT----------------------------GTTTTGCCGGTCGTACCCGGTTTTT-ATTCGCA-T----TTTA-----T |
| droEle1 |
scf7180000490967:982638-982695 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTCACGGCGC---AGCACAAGTGAGTAC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droRho1 |
scf7180000779916:74756-74813 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCAGCTCACGGCGC---AGCACAAGTGAGTAC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droFic1 |
scf7180000454104:1086552-1086609 - |
|
AACACCACACACAGTATTGAGGTGCCAT---CGCAGCTGGCGGCGC---AGCACAAGTGAGTAC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000302706:1990376-1990515 + |
|
ACCACCACACACAGTATTGAGGTGCCAC---CGCCGCTGACGCCGC---AGCACAAGTGAGTACCACTC--------------------------AGAT-------TCTACATGTATT----------T-------TA-------------AAGATGAAAGTATAAA---------------------GAAAACAGGGAA--------------------------------TATTAAAAAATAATCATAAGACTTTA-------------------------------------------------------------------------------------------------------------------------------------TGGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droAna3 |
scaffold_13340:6316101-6316181 + |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCCGCTGACGCCGC---AGCACAAGTGAGTAGATGTA--------------------------GATA-------CTTTTTATAAGA----------TA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droBip1 |
scf7180000396708:2527813-2527898 - |
|
ACCACCACACACAGTATTGAGGTGCCAT---CGCCGCTGACGCCGC---AACACAAGTGAGTAGATACC--------------------------ATA------------------------------TAGCA-----------------------------------------------------TA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAGATAGTAGTC--------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp5 |
2:5218152-5218211 - |
|
ACCACCACCCACAGTATTGAGGTGCTGC---CACCTCTGACGGCACAACATCACAAGTGAGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_19:928009-928193 - |
|
ACCACCACCCACAGTATTGAGGTGCTGC---CACCTCTGACGGCACAACAGCACAAGTGAGTAGACCCT--------------------------AGAC----------TCTAGACTC----------TAGTC-TGTA-------------GATTGTAGA--------------------------------------------------------------------------------------------------TGATTGATCCCCCCCACTACACG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AC--------CCACTATATTATATGTGT-GAATGAATGGTTTGGTTTGCCTGCCGGTTGTACCCGT-TTGG-AATC------------------- |
| droWil2 |
scf2_1100000004943:16312288-16312336 - |
|
--TACCACACACAGTATTGAGGTGCCAT---------TGACGCCAC---AGCACAAGTAAGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_13047:10764737-10765023 - |
|
--TACCACACACAGTATTGAGGTGCCATTGACGCCACTGACGCCGC---CGAACAAGTGAGTAAACCAGATTAGATTTGAATGCCCG---TTCAACA-------TACCCTC----------------------------------------------------------------------------------------------GCACCAATAAACAAACTCTAGATTTTC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTA------------TCAA-GTGACAGTGAGGCAGATAATAATCAATGCATATATACTTAGAATAAACGCATTTTCACATAAATTAATTTATTGCCAATTATAAAACAATTATG-----------------TATGTTACATAGATCGTTTGGATAGTT---------TATAGGGTGTTC-TTTCGCCGAGCTGTTTATATTTG |
| droMoj3 |
scaffold_6540:31028475-31028572 + |
|
--TACCACACACAGTATTGAGGTGCCAT---TGACGCAGACGCCGC---CGAACAAGTGAGTAGCAC-------------------GACTTTGCACA-------TACCCTCTTC--------------TAGTA-----------------------------------------------------CA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAGGCAATGG----------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_14624:541286-541342 - |
|
AC---CACACACAGTATTGAGGTGCCATTGACGCCGCTGACGCCGC---CAAATAAGTGAGTA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |